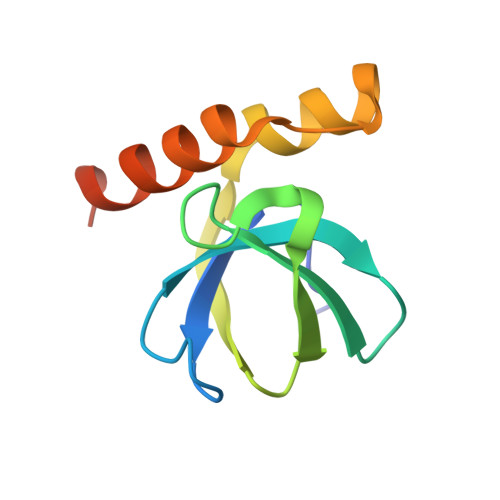
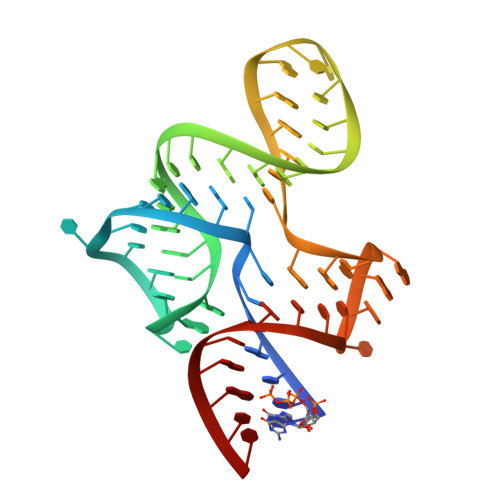
Structure of a bacterial OapB protein with its OLE RNA target gives insights into the architecture of the OLE ribonucleoprotein complex.
Yang, Y., Harris, K.A., Widner, D.L., Breaker, R.R.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 33619097
- DOI: https://doi.org/10.1073/pnas.2020393118
- Primary Citation of Related Structures:
7K9B, 7K9C, 7K9D, 7K9E, 7KKV - PubMed Abstract:
The OLE (ornate, large, and extremophilic) RNA class is one of the most complex and well-conserved bacterial noncoding RNAs known to exist. This RNA is known to be important for bacterial responses to stress caused by short-chain alcohols, cold, and elevated Mg 2+ concentrations. These biological functions have been shown to require the formation of a ribonucleoprotein (RNP) complex including at least two protein partners: OLE-associated protein A (OapA) and OLE-associated protein B (OapB). OapB directly binds OLE RNA with high-affinity and specificity and is believed to assist in assembling the functional OLE RNP complex. To provide the atomic details of OapB-OLE RNA interaction and to potentially reveal previously uncharacterized protein-RNA interfaces, we determined the structure of OapB from Bacillus halodurans alone and in complex with an OLE RNA fragment at resolutions of 1.0 Å and 2.0 Å, respectively. The structure of OapB exhibits a K-shaped overall architecture wherein its conserved KOW motif and additional unique structural elements of OapB form a bipartite RNA-binding surface that docks to the P13 hairpin and P12.2 helix of OLE RNA. These high-resolution structures elucidate the molecular contacts used by OapB to form a stable RNP complex and explain the high conservation of sequences and structural features at the OapB-OLE RNA-binding interface. These findings provide insight into the role of OapB in the assembly and biological function of OLE RNP complex and can guide the exploration of additional possible OLE RNA-binding interactions present in OapB.
- Department of Molecular, Cellular and Developmental Biology, Yale University, New Haven, CT 06520-8103; y.yang@yale.edu ronald.breaker@yale.edu.
Organizational Affiliation: