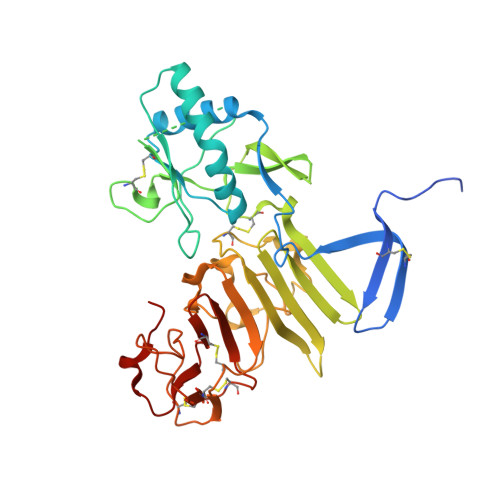
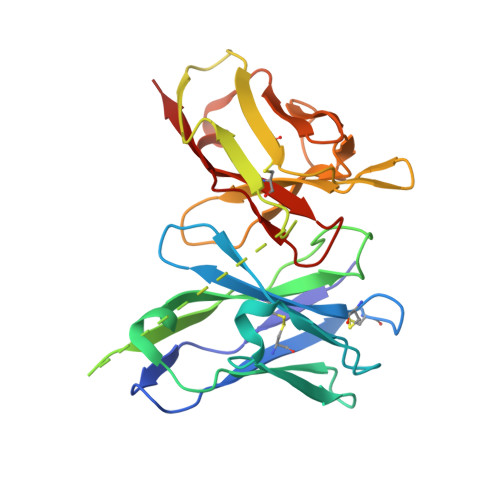
Structural basis for antibody inhibition of flavivirus NS1-triggered endothelial dysfunction.
Biering, S.B., Akey, D.L., Wong, M.P., Brown, W.C., Lo, N.T.N., Puerta-Guardo, H., Tramontini Gomes de Sousa, F., Wang, C., Konwerski, J.R., Espinosa, D.A., Bockhaus, N.J., Glasner, D.R., Li, J., Blanc, S.F., Juan, E.Y., Elledge, S.J., Mina, M.J., Beatty, P.R., Smith, J.L., Harris, E.(2021) Science 371: 194-200
- PubMed: 33414220
- DOI: https://doi.org/10.1126/science.abc0476
- Primary Citation of Related Structures:
6WEQ, 6WER, 7K93 - PubMed Abstract:
Medically important flaviviruses cause diverse disease pathologies and collectively are responsible for a major global disease burden. A contributing factor to pathogenesis is secreted flavivirus nonstructural protein 1 (NS1). Despite demonstrated protection by NS1-specific antibodies against lethal flavivirus challenge, the structural and mechanistic basis remains unknown. Here, we present three crystal structures of full-length dengue virus NS1 complexed with a flavivirus-cross-reactive, NS1-specific monoclonal antibody, 2B7, at resolutions between 2.89 and 3.96 angstroms. These structures reveal a protective mechanism by which two domains of NS1 are antagonized simultaneously. The NS1 wing domain mediates cell binding, whereas the β-ladder triggers downstream events, both of which are required for dengue, Zika, and West Nile virus NS1-mediated endothelial dysfunction. These observations provide a mechanistic explanation for 2B7 protection against NS1-induced pathology and demonstrate the potential of one antibody to treat infections by multiple flaviviruses.
- Division of Infectious Diseases and Vaccinology, School of Public Health, University of California, Berkeley, Berkeley, CA 94720-3370, USA.
Organizational Affiliation: