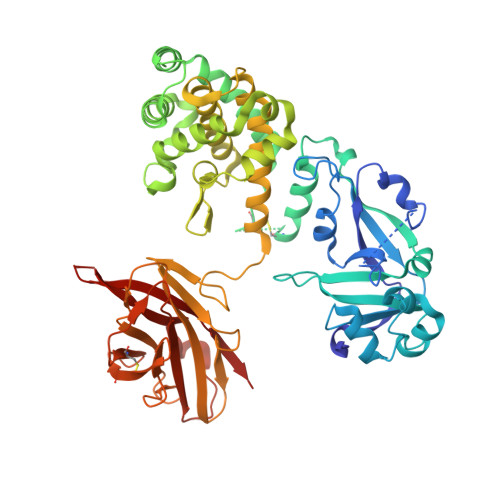
Structure of the Diphtheria Toxin at Acidic pH: Implications for the Conformational Switching of the Translocation Domain.
Rodnin, M.V., Kashipathy, M.M., Kyrychenko, A., Battaile, K.P., Lovell, S., Ladokhin, A.S.(2020) Toxins (Basel) 12
- PubMed: 33171806
- DOI: https://doi.org/10.3390/toxins12110704
- Primary Citation of Related Structures:
7K7B, 7K7C, 7K7D, 7K7E - PubMed Abstract:
Diphtheria toxin, an exotoxin secreted by Corynebacterium that causes disease in humans by inhibiting protein synthesis, enters the cell via receptor-mediated endocytosis. The subsequent endosomal acidification triggers a series of conformational changes, resulting in the refolding and membrane insertion of the translocation (T-)domain and ultimately leading to the translocation of the catalytic domain into the cytoplasm. Here, we use X-ray crystallography along with circular dichroism and fluorescence spectroscopy to gain insight into the mechanism of the early stages of pH-dependent conformational transition. For the first time, we present the high-resolution structure of the diphtheria toxin at a mildly acidic pH (5-6) and compare it to the structure at neutral pH (7). We demonstrate that neither catalytic nor receptor-binding domains change their structure upon this acidification, while the T-domain undergoes a conformational change that results in the unfolding of the TH2-3 helices. Surprisingly, the TH1 helix maintains its conformation in the crystal of the full-length toxin even at pH 5. This contrasts with the evidence from the new and previously published data, obtained by spectroscopic measurements and molecular dynamics computer simulations, which indicate the refolding of TH1 upon the acidification of the isolated T-domain. The overall results imply that the membrane interactions of the T-domain are critical in ensuring the proper conformational changes required for the preparation of the diphtheria toxin for the cellular entry.
- Department of Biochemistry and Molecular Biology, University of Kansas Medical Center, Kansas City, KS 66160, USA.
Organizational Affiliation: