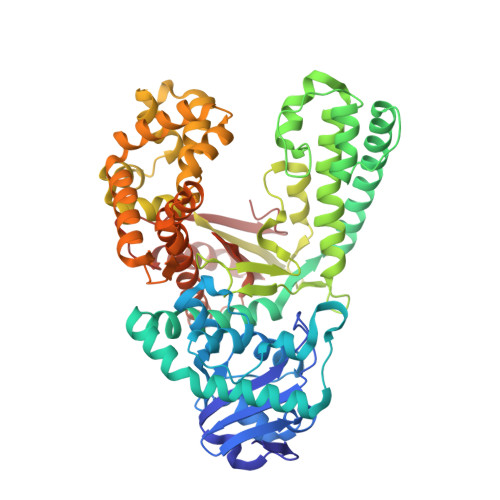
Following replicative DNA synthesis by time-resolved X-ray crystallography.
Chim, N., Meza, R.A., Trinh, A.M., Yang, K., Chaput, J.C.(2021) Nat Commun 12: 2641-2641
- PubMed: 33976175
- DOI: https://doi.org/10.1038/s41467-021-22937-z
- Primary Citation of Related Structures:
7K5O, 7K5P, 7K5Q, 7K5R, 7K5S, 7K5T, 7K5U - PubMed Abstract:
The mechanism of DNA synthesis has been inferred from static structures, but the absence of temporal information raises longstanding questions about the order of events in one of life's most central processes. Here we follow the reaction pathway of a replicative DNA polymerase using time-resolved X-ray crystallography to elucidate the order and transition between intermediates. In contrast to the canonical model, the structural changes observed in the time-lapsed images reveal a catalytic cycle in which translocation precedes catalysis. The translocation step appears to follow a push-pull mechanism where the O-O1 loop of the finger subdomain acts as a pawl to facilitate unidirectional movement along the template with conserved tyrosine residues 714 and 719 functioning as tandem gatekeepers of DNA synthesis. The structures capture the precise order of critical events that may be a general feature of enzymatic catalysis among replicative DNA polymerases.
- Department of Pharmaceutical Sciences, University of California, Irvine, CA, USA.
Organizational Affiliation: