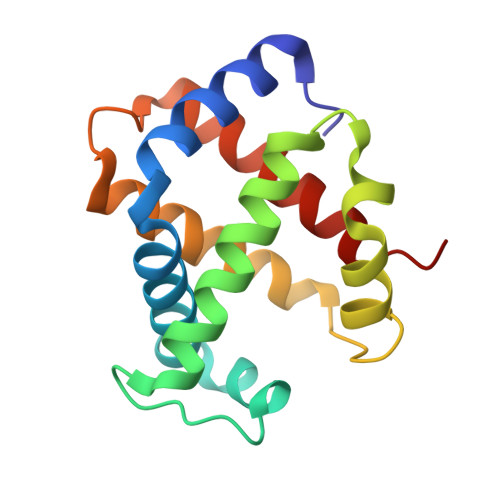
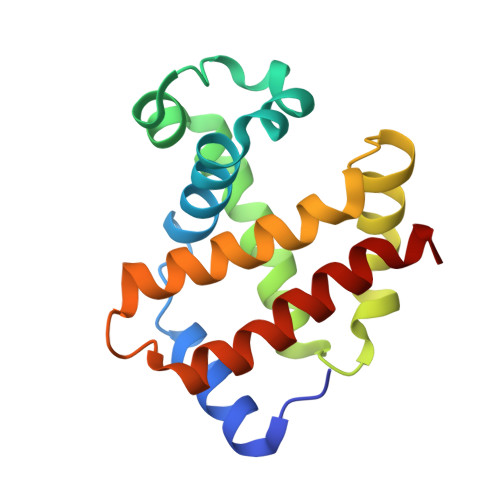
MetAP2 inhibition modifies hemoglobin S to delay polymerization and improves blood flow in sickle cell disease.
Demers, M., Sturtevant, S., Guertin, K.R., Gupta, D., Desai, K., Vieira, B.F., Li, W., Hicks, A., Ismail, A., Goncalves, B.P., Di Caprio, G., Schonbrun, E., Hansen, S., Musayev, F.N., Safo, M.K., Wood, D.K., Higgins, J.M., Light, D.R.(2021) Blood Adv 5: 1388-1402
- PubMed: 33661300
- DOI: https://doi.org/10.1182/bloodadvances.2020003670
- Primary Citation of Related Structures:
7K4M - PubMed Abstract:
Sickle cell disease (SCD) is associated with hemolysis, vascular inflammation, and organ damage. Affected patients experience chronic painful vaso-occlusive events requiring hospitalization. Hypoxia-induced polymerization of sickle hemoglobin S (HbS) contributes to sickling of red blood cells (RBCs) and disease pathophysiology. Dilution of HbS with nonsickling hemoglobin or hemoglobin with increased oxygen affinity, such as fetal hemoglobin or HbS bound to aromatic aldehydes, is clinically beneficial in decreasing polymerization. We investigated a novel alternate approach to modify HbS and decrease polymerization by inhibiting methionine aminopeptidase 2 (MetAP2), which cleaves the initiator methionine (iMet) from Val1 of α-globin and βS-globin. Kinetic studies with MetAP2 show that βS-globin is a fivefold better substrate than α-globin. Knockdown of MetAP2 in human umbilical cord blood-derived erythroid progenitor 2 cells shows more extensive modification of α-globin than β-globin, consistent with kinetic data. Treatment of human erythroid cells in vitro or Townes SCD mice in vivo with selective MetAP2 inhibitors extensively modifies both globins with N-terminal iMet and acetylated iMet. HbS modification by MetAP2 inhibition increases oxygen affinity, as measured by decreased oxygen tension at which hemoglobin is 50% saturated. Acetyl-iMet modification on βS-globin delays HbS polymerization under hypoxia. MetAP2 inhibitor-treated Townes mice reach 50% total HbS modification, significantly increasing the affinity of RBCs for oxygen, increasing whole blood single-cell RBC oxygen saturation, and decreasing fractional flow velocity losses in blood rheology under decreased oxygen pressures. Crystal structures of modified HbS variants show stabilization of the nonpolymerizing high O2-affinity R2 state, explaining modified HbS antisickling activity. Further study of MetAP2 inhibition as a potential therapeutic target for SCD is warranted.
- Department of Rare Blood Disorders, Sanofi, Waltham, MA.
Organizational Affiliation: