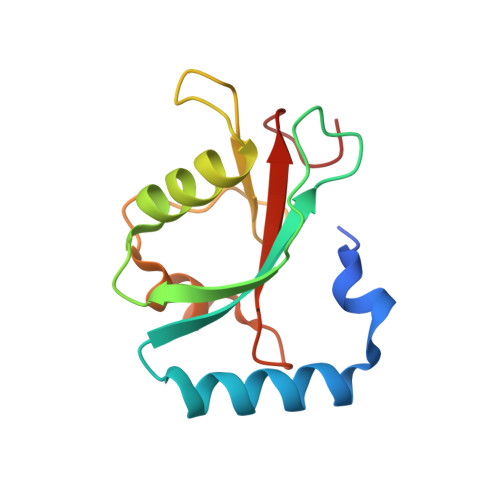
Insights on autophagosome-lysosome tethering from structural and biochemical characterization of human autophagy factor EPG5.
Nam, S.E., Cheung, Y.W.S., Nguyen, T.N., Gong, M., Chan, S., Lazarou, M., Yip, C.K.(2021) Commun Biol 4: 291-291
- PubMed: 33674710
- DOI: https://doi.org/10.1038/s42003-021-01830-x
- Primary Citation of Related Structures:
7JHX - PubMed Abstract:
Pivotal to the maintenance of cellular homeostasis, macroautophagy (hereafter autophagy) is an evolutionarily conserved degradation system that involves sequestration of cytoplasmic material into the double-membrane autophagosome and targeting of this transport vesicle to the lysosome/late endosome for degradation. EPG5 is a large-sized metazoan protein proposed to serve as a tethering factor to enforce autophagosome-lysosome/late endosome fusion specificity, and its deficiency causes a severe multisystem disorder known as Vici syndrome. Here, we show that human EPG5 (hEPG5) adopts an extended "shepherd's staff" architecture. We find that hEPG5 binds preferentially to members of the GABARAP subfamily of human ATG8 proteins critical to autophagosome-lysosome fusion. The hEPG5-GABARAPs interaction, which is mediated by tandem LIR motifs that exhibit differential affinities, is required for hEPG5 recruitment to mitochondria during PINK1/Parkin-dependent mitophagy. Lastly, we find that the Vici syndrome mutation Gln336Arg does not affect the hEPG5's overall stability nor its ability to engage in interaction with the GABARAPs. Collectively, results from our studies reveal new insights into how hEPG5 recognizes mature autophagosome and establish a platform for examining the molecular effects of Vici syndrome disease mutations on hEPG5.
- Life Sciences Institute, Department of Biochemistry and Molecular Biology, The University of British Columbia, Vancouver, BC, Canada.
Organizational Affiliation: