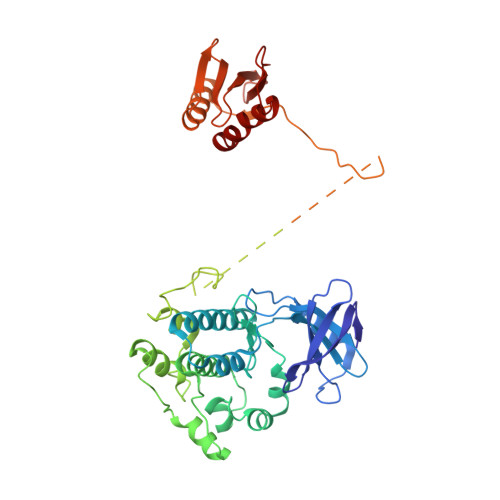
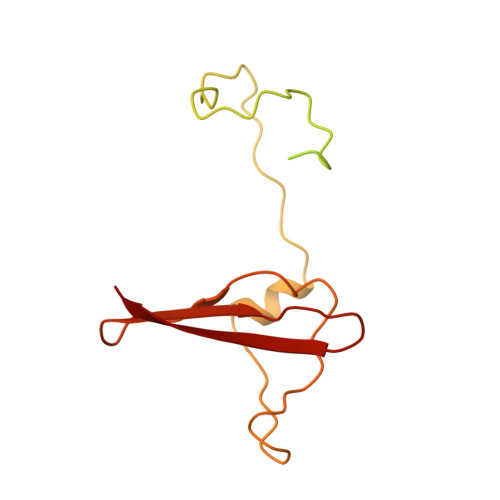
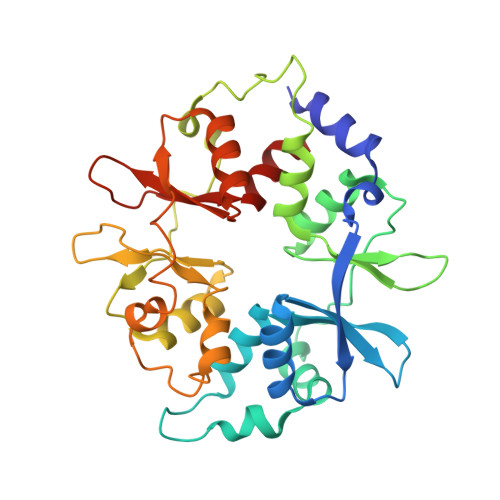
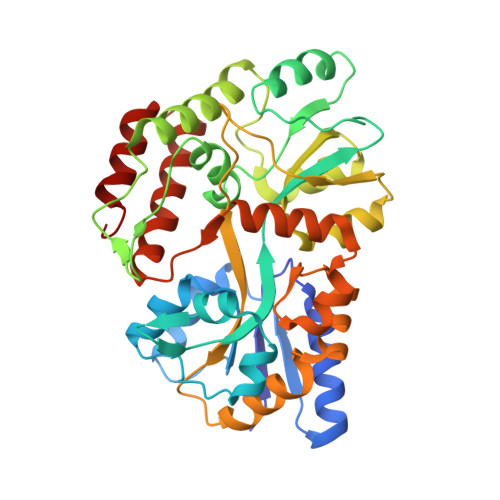
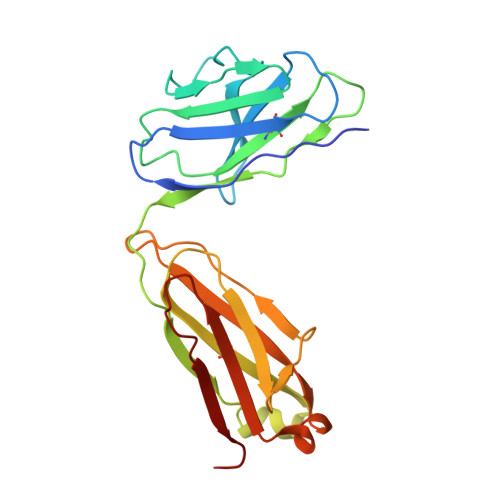
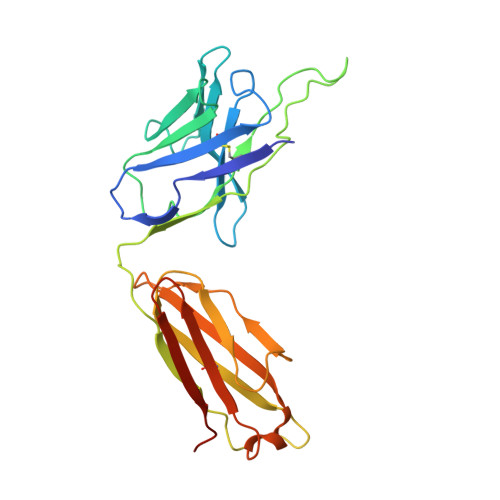
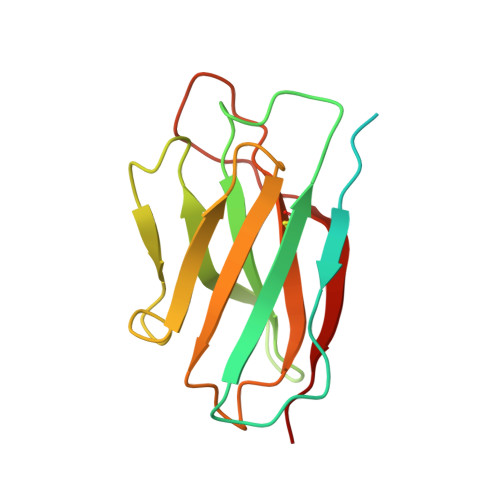
Structure of an AMPK complex in an inactive, ATP-bound state.
Yan, Y., Mukherjee, S., Harikumar, K.G., Strutzenberg, T.S., Zhou, X.E., Suino-Powell, K., Xu, T.H., Sheldon, R.D., Lamp, J., Brunzelle, J.S., Radziwon, K., Ellis, A., Novick, S.J., Vega, I.E., Jones, R.G., Miller, L.J., Xu, H.E., Griffin, P.R., Kossiakoff, A.A., Melcher, K.(2021) Science 373: 413-419
- PubMed: 34437114
- DOI: https://doi.org/10.1126/science.abe7565
- Primary Citation of Related Structures:
7JHG, 7JHH, 7JIJ, 7M74 - PubMed Abstract:
Adenosine monophosphate (AMP)-activated protein kinase (AMPK) regulates metabolism in response to the cellular energy states. Under energy stress, AMP stabilizes the active AMPK conformation, in which the kinase activation loop (AL) is protected from protein phosphatases, thus keeping the AL in its active, phosphorylated state. At low AMP:ATP (adenosine triphosphate) ratios, ATP inhibits AMPK by increasing AL dynamics and accessibility. We developed conformation-specific antibodies to trap ATP-bound AMPK in a fully inactive, dynamic state and determined its structure at 3.5-angstrom resolution using cryo-electron microscopy. A 180° rotation and 100-angstrom displacement of the kinase domain fully exposes the AL. On the basis of the structure and supporting biophysical data, we propose a multistep mechanism explaining how adenine nucleotides and pharmacological agonists modulate AMPK activity by altering AL phosphorylation and accessibility.
- Department of Structural Biology, Van Andel Institute, Grand Rapids, MI 49503, USA.
Organizational Affiliation: