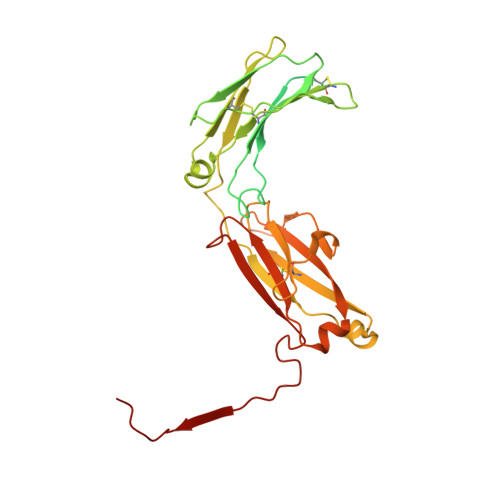
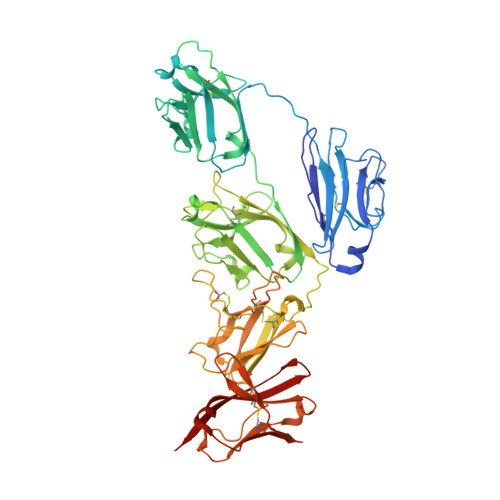
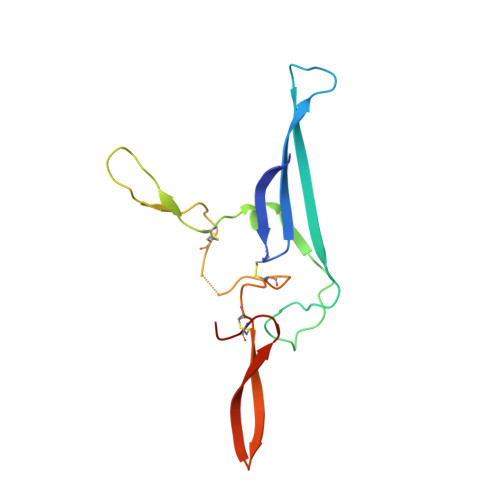
The structures of Secretory and dimeric Immunoglobulin A.
Kumar Bharathkar, S., Parker, B.W., Malyutin, A.G., Haloi, N., Huey-Tubman, K.E., Tajkhorshid, E., Stadtmueller, B.(2020) Elife 9
- PubMed: 33107820
- DOI: https://doi.org/10.7554/eLife.56098
- Primary Citation of Related Structures:
7JG1, 7JG2 - PubMed Abstract:
Secretory (S) Immunoglobulin (Ig) A is the predominant mucosal antibody, which binds pathogens and commensal microbes. SIgA is a polymeric antibody, typically containing two copies of IgA that assemble with one joining-chain (JC) to form dimeric (d) IgA that is bound by the polymeric Ig-receptor ectodomain, called secretory component (SC). Here, we report the cryo-electron microscopy structures of murine SIgA and dIgA. Structures reveal two IgAs conjoined through four heavy-chain tailpieces and the JC that together form a β-sandwich-like fold. The two IgAs are bent and tilted with respect to each other, forming distinct concave and convex surfaces. In SIgA, SC is bound to one face, asymmetrically contacting both IgAs and JC. The bent and tilted arrangement of complex components limits the possible positions of both sets of antigen-binding fragments (Fabs) and preserves steric accessibility to receptor-binding sites, likely influencing antigen binding and effector functions.
- Department of Biochemistry, University of Illinois Urbana-Champaign, Urbana, United States.
Organizational Affiliation: