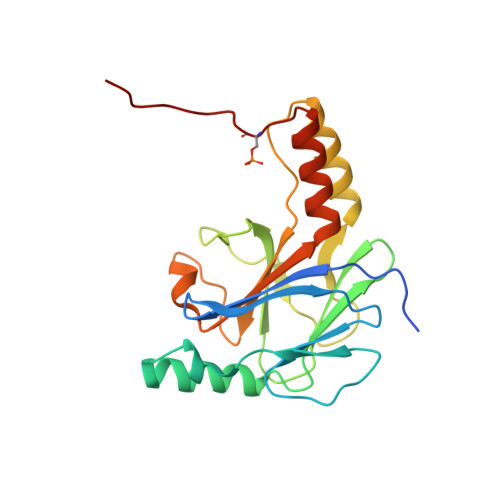
The Structural Basis of IRF-3 Activation upon Phosphorylation.
Jing, T., Zhao, B., Xu, P., Gao, X., Chi, L., Han, H., Sankaran, B., Li, P.(2020) J Immunol 205: 1886-1896
- PubMed: 32826280
- DOI: https://doi.org/10.4049/jimmunol.2000026
- Primary Citation of Related Structures:
7JFL, 7JFM - PubMed Abstract:
The innate immune system is the first line of defense against bacterial and viral infections. The recognition of pathogen-associated molecular patterns by the RIG-I-like receptors, TLRs, and cGAS leads to the induction of IFN-I by activating the transcription factor IRF-3. Although the mechanism of IRF-3 activation has been extensively studied, the structural basis of IRF-3 activation upon phosphorylation is not fully understood. In this study, we determined the crystal structures of phosphorylated human and mouse IRF-3 bound to CREB-binding protein (CBP), which reveal that phosphorylated IRF-3 forms a dimer via pSer 386 (pSer 379 in mouse IRF-3) and a downstream p L x IS motif. Size-exclusion chromatography and cell-based studies show that mutations of key residues interacting with pSer 386 severely impair IRF-3 activation and IFN-β induction. By contrast, phosphorylation of Ser 396 within the p L x IS motif of human IRF-3 only plays a moderate role in IRF-3 activation. The mouse IRF-3/CBP complex structure reveals that the mechanism of mouse IRF-3 activation is similar but distinct from human IRF-3. These structural and functional studies reveal the detailed mechanism of IRF-3 activation upon phosphorylation.
- Department of Biochemistry and Biophysics, Texas A&M University, College Station, TX 77843.
Organizational Affiliation: