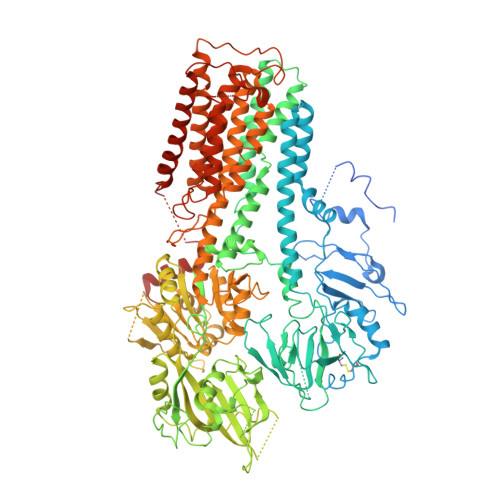
Cryo-EM structures and transport mechanism of human P5B type ATPase ATP13A2.
Chen, X., Zhou, M., Zhang, S., Yin, J., Zhang, P., Xuan, X., Wang, P., Liu, Z., Zhou, B., Yang, M.(2021) Cell Discov 7: 106-106
- PubMed: 34728622
- DOI: https://doi.org/10.1038/s41421-021-00334-6
- Primary Citation of Related Structures:
7FJM, 7FJP, 7FJQ - PubMed Abstract:
Polyamines are important polycations that play critical roles in mammalian cells. ATP13A2 belongs to the orphan P5B adenosine triphosphatases (ATPase) family and has been established as a lysosomal polyamine exporter to maintain the normal function of lysosomes and mitochondria. Previous studies have reported that several human neurodegenerative disorders are related to mutations in the ATP13A2 gene. However, the transport mechanism of ATP13A2 in the lysosome remains unclear. Here, we report the cryo-electron microscopy (cryo-EM) structures of three distinct intermediates of the human ATP13A2, revealing key insights into the spermine (SPM) transport cycle in the lysosome. The transmembrane domain serves as a substrate binding site and the C-terminal domain is essential for protein stability and may play a regulatory role. These findings advance our understanding of the polyamine transport mechanism, the lipid-associated regulation, and the disease-associated mutants of ATP13A2.
- Ministry of Education Key Laboratory of Protein Science, Tsinghua-Peking Center for Life Sciences, Beijing Advanced Innovation Center for Structural Biology, School of Life Sciences, Tsinghua University, Beijing, China.
Organizational Affiliation: