Structural basis of the IL-1 receptor TIR domain-mediated IL-1 signaling
Zhou, J., Xiao, Y., Ren, Y., Ge, J., Wang, X.(2022) iScience 25: 104508
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Interleukin-18 receptor accessory protein | 166 | Homo sapiens | Mutation(s): 0 Gene Names: IL18RAP EC: 3.2.2.6 | 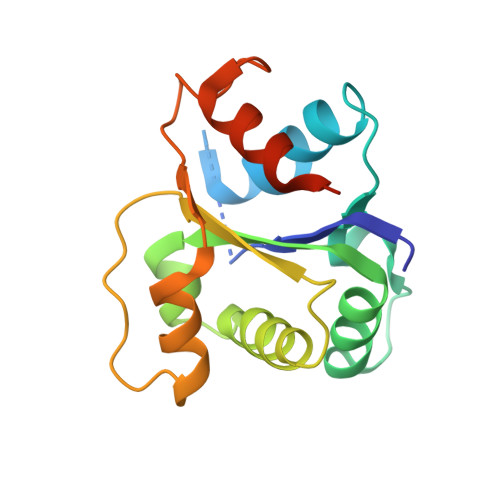 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for O95256 (Homo sapiens) Explore O95256 Go to UniProtKB: O95256 | |||||
PHAROS: O95256 GTEx: ENSG00000115607 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | O95256 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 39.76 | α = 90 |
| b = 161.074 | β = 112.44 |
| c = 52.164 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| PHASER | phasing |