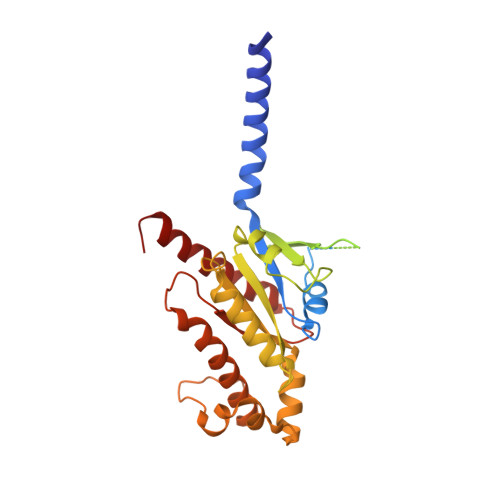
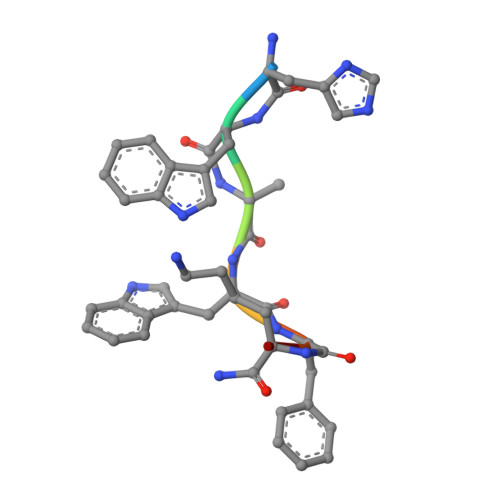
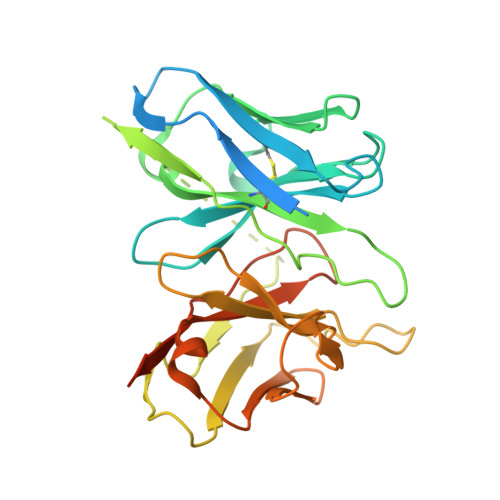
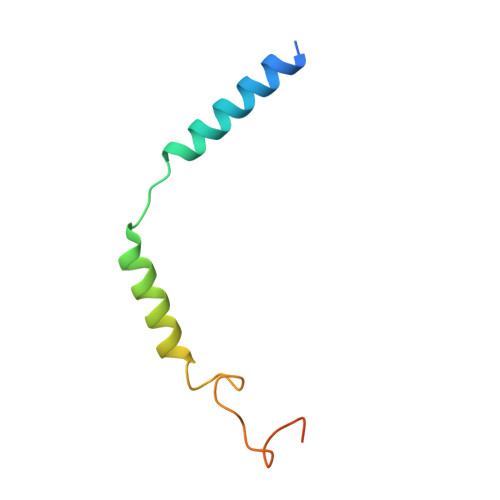
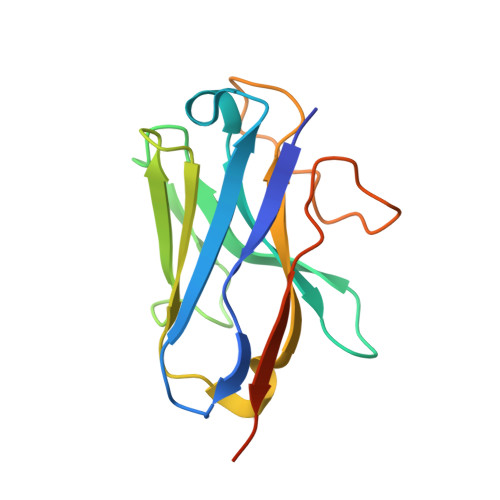
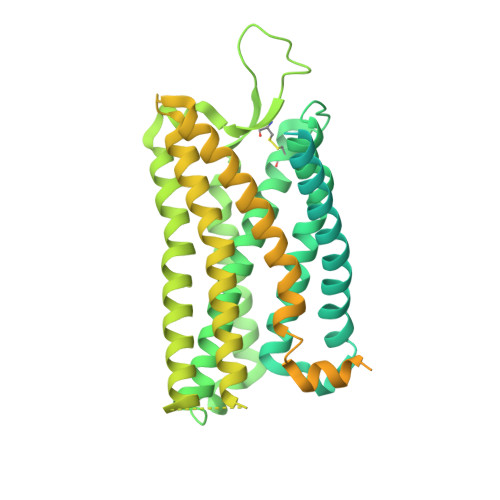
Molecular recognition of an acyl-peptide hormone and activation of ghrelin receptor.
Wang, Y., Guo, S., Zhuang, Y., Yun, Y., Xu, P., He, X., Guo, J., Yin, W., Xu, H.E., Xie, X., Jiang, Y.(2021) Nat Commun 12: 5064-5064
- PubMed: 34417468
- DOI: https://doi.org/10.1038/s41467-021-25364-2
- Primary Citation of Related Structures:
7F9Y, 7F9Z - PubMed Abstract:
Ghrelin, also called "the hunger hormone", is a gastric peptide hormone that regulates food intake, body weight, as well as taste sensation, reward, cognition, learning and memory. One unique feature of ghrelin is its acylation, primarily with an octanoic acid, which is essential for its binding and activation of the ghrelin receptor, a G protein-coupled receptor. The multifaceted roles of ghrelin make ghrelin receptor a highly attractive drug target for growth retardation, obesity, and metabolic disorders. Here we present two cryo-electron microscopy structures of G q -coupled ghrelin receptor bound to ghrelin and a synthetic agonist, GHRP-6. Analysis of these two structures reveals a unique binding pocket for the octanoyl group, which guides the correct positioning of the peptide to initiate the receptor activation. Together with mutational and functional data, our structures define the rules for recognition of the acylated peptide hormone and activation of ghrelin receptor, and provide structural templates to facilitate drug design targeting ghrelin receptor.
- CAS Key Laboratory of Receptor Research, Center for Structure and Function of Drug Targets, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, Shanghai, China.
Organizational Affiliation: