TgPRS with double inhibitors
Malhotra, N., Manickam, Y., Sharma, A.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Prolyl-tRNA synthetase (ProRS) | 500 | Toxoplasma gondii | Mutation(s): 0 Gene Names: TGRH88_057780 EC: 6.1.1.15 | 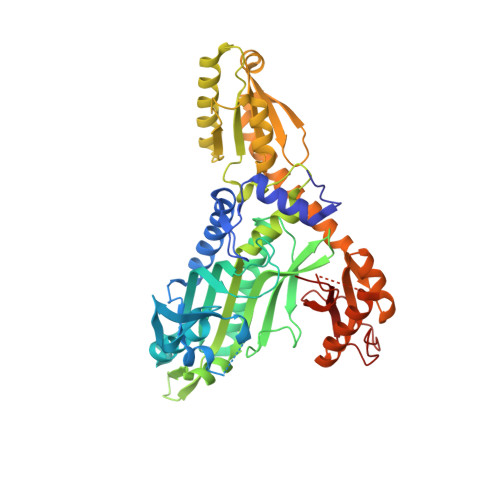 | |
UniProt | |||||
Find proteins for A0A7J6JUK2 (Toxoplasma gondii) Explore A0A7J6JUK2 Go to UniProtKB: A0A7J6JUK2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A0A7J6JUK2 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 8 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| 1TI (Subject of Investigation/LOI) Query on 1TI | C [auth A], Q [auth B] | 4-[(3S)-3-cyano-3-(1-methylcyclopropyl)-2-oxidanylidene-pyrrolidin-1-yl]-N-[[3-fluoranyl-5-(1-methylpyrazol-4-yl)phenyl]methyl]-6-methyl-pyridine-2-carboxamide C27 H27 F N6 O2 PFICVJYRZDMEIX-MHZLTWQESA-N |  | ||
| HFG Query on HFG | D [auth A], R [auth B] | 7-bromo-6-chloro-3-{3-[(2R,3S)-3-hydroxypiperidin-2-yl]-2-oxopropyl}quinazolin-4(3H)-one C16 H17 Br Cl N3 O3 LVASCWIMLIKXLA-CABCVRRESA-N |  | ||
| MES Query on MES | E [auth A] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| IOD Query on IOD | H [auth A] I [auth A] K [auth A] L [auth A] P [auth B] | IODIDE ION I XMBWDFGMSWQBCA-UHFFFAOYSA-M |  | ||
| GOL Query on GOL | S [auth B] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| BR Query on BR | AA [auth B] BA [auth B] G [auth A] J [auth A] M [auth A] | BROMIDE ION Br CPELXLSAUQHCOX-UHFFFAOYSA-M |  | ||
| BME Query on BME | F [auth A] | BETA-MERCAPTOETHANOL C2 H6 O S DGVVWUTYPXICAM-UHFFFAOYSA-N |  | ||
| F Query on F | N [auth A], O [auth A], Y [auth B] | FLUORIDE ION F KRHYYFGTRYWZRS-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 77.067 | α = 90 |
| b = 82.981 | β = 103.08 |
| c = 105.521 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| autoPROC | data reduction |
| autoPROC | data scaling |
| PHASER | phasing |