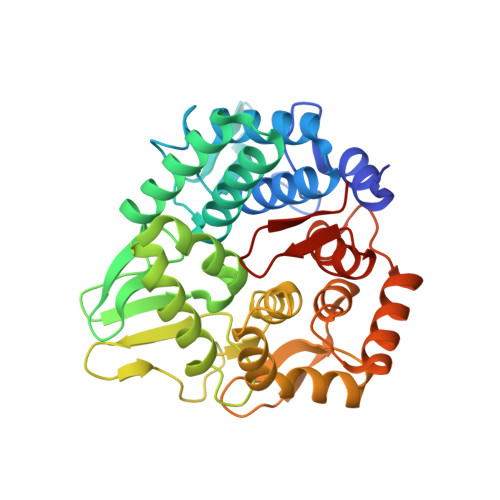
Structural snapshot of a glycoside hydrolase family 8 endo-beta-1,4-glucanase capturing the state after cleavage of the scissile bond.
Fujiwara, T., Fujishima, A., Nakamura, Y., Tajima, K., Yao, M.(2022) Acta Crystallogr D Biol Crystallogr 78: 228-237