Structural basis of human PDZD8-Rab7 interaction for the ER-late endosome tethering.
Khan, H., Chen, L., Tan, L., Im, Y.J.(2021) Sci Rep 11: 18859-18859
- PubMed: 34552186
- DOI: https://doi.org/10.1038/s41598-021-98419-5
- Primary Citation of Related Structures:
7F6J - PubMed Abstract:
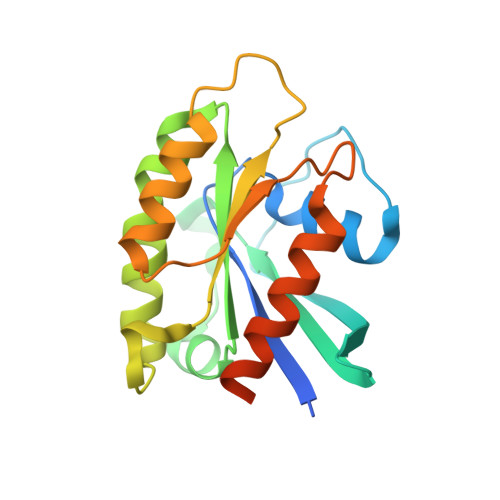
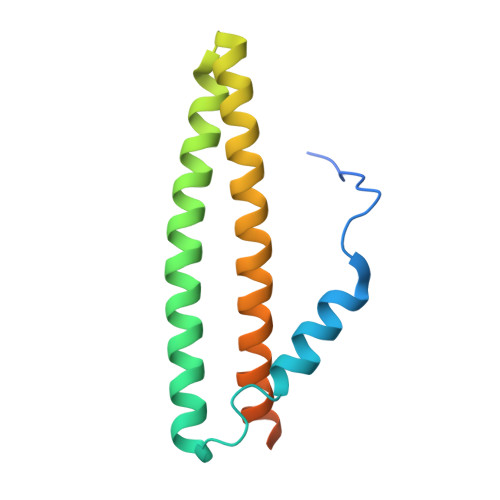
The membrane contact sites (MCSs) between the ER and late endosomes (LEs) are essential for the regulation of endosomal protein sorting, dynamics, and motility. PDZD8 is an ER transmembrane protein containing a Synaptotagmin-like Mitochondrial lipid-binding Proteins (SMP) domain. PDZD8 tethers the ER to late endosomes and lysosomes by associating its C-terminal coiled-coil (CC) with the LE Rab7. To identify the structural determinants for the PDZD8-Rab7 interaction, we determined the crystal structure of the human PDZD8 CC domain in complex with the GTP-bound form of Rab7. The PDZD8 CC contains one short helix and the two helices forming an antiparallel coiled-coil. Two Rab7 molecules bind to the opposite sides of the PDZD8 CC in a 2:1 ratio. The switch I/II and interswitch regions of the GTP-loaded Rab7 form the binding interfaces, which correlates with the GTP-dependent interaction of PDZD8 and Rab7. Analysis of the protein interaction by isothermal titration calorimetry confirms that two Rab7 molecules bind the PDZD8 CC in a GTP-dependent manner. The structural model of the PDZD8 CC-Rab7 complex correlates with the recruitment of PDZD8 at the LE-ER interface and its role in lipid transport and regulation.
- College of Pharmacy, Chonnam National University, Gwangju, 61186, Republic of Korea.
Organizational Affiliation: