The development and characterization of a long acting anti-thrombotic von Willebrand factor (VWF) aptamer
Zhu, S., Gilbert, C.J.(2020) J Thromb Haemost 18: 1113-1123
- PubMed: 32011054
- DOI: https://doi.org/10.1111/jth.14755
- Primary Citation of Related Structures:
7F49 - PubMed Abstract:
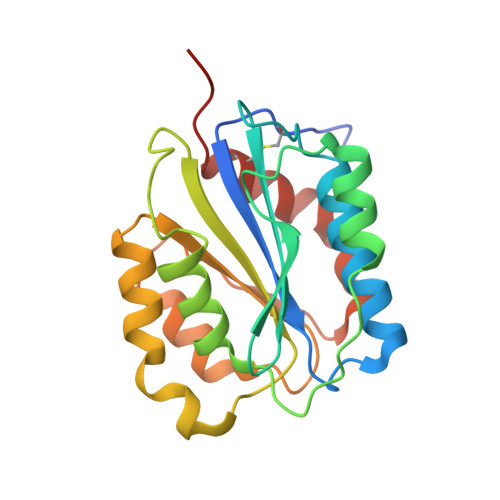
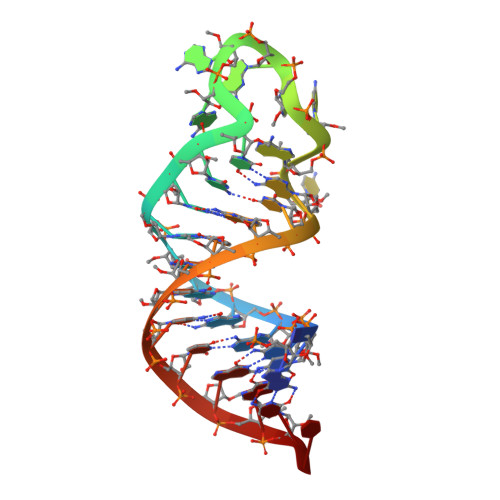
Thrombus formation involves coagulation proteins and platelets. The latter, referred to as platelet-mediated thrombogenesis, is predominant in arterial circulation. Platelet thrombogenesis follows vascular injury when extracellular von Willebrand factor (VWF) binds via its A3 domain to exposed collagen, and the free VWF A1 domain binds to platelet glycoprotein Ib (GPIb). To characterize the antiplatelet/antithrombotic activity of the pegylated VWF antagonist aptamer BT200 and identify the aptamer VWF binding site. BT100 is an optimized aptamer synthesized by solid-phase chemistry and pegylated (BT200) by standard conjugation chemistry. The affinity of BT200 for purified human VWF was evaluated as was VWF inhibition in monkey and human plasma. Efficacy of BT200 was assessed in the monkey FeCl 3 femoral artery thrombosis model. BT200 bound human VWF at an EC 50 of 5.0 nmol/L and inhibited VWF A1 domain activity in monkey and human plasma with mean IC 50 values of 183 and 70 nmol/L. BT200 administration to cynomolgus monkeys caused a time-dependent and dose-dependent effect on VWF A1 domain activity and inhibited platelet function as measured by collagen adenosine diphosphate closure time in the platelet function analyzer. BT200 demonstrated a bioavailability of ≥77% and exhibited a half-life of >100 hours after subcutaneous injection. The treatment effectively prevented arterial occlusion in an FeCl 3 -induced thrombosis model in monkeys. BT200 has shown promising inhibition of human VWF in vitro and prevented arterial occlusion in non-human primates. These data including a long half-life after subcutaneous injections provide a strong rationale for ongoing clinical development of BT200.
- Guardian Therapeutics Inc, Lexington, Massachusetts, USA.
Organizational Affiliation: