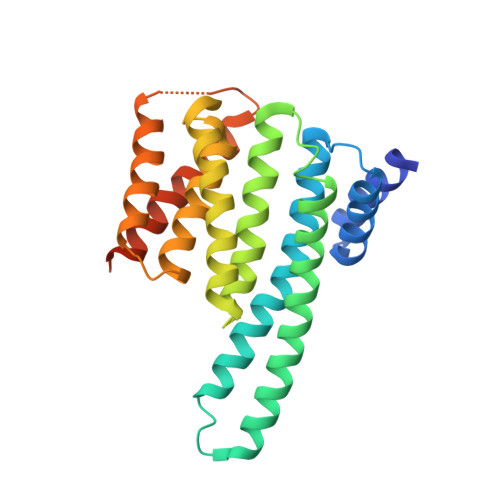
LGI1-ADAM22 levels regulate seizure thresholds through 14-3-3 in mice
Yokoi, N., Fukata, Y., Okatsu, K., Yamagata, A., Yan, L., Sanbo, M., Miyazaki, Y., Goto, T., Abe, M., Natsume, R., Aiba, A., Sakimura, K., Meijer, D., Hirabayashi, M., Fukai, S., Fukata, M.(2021) Cell Rep