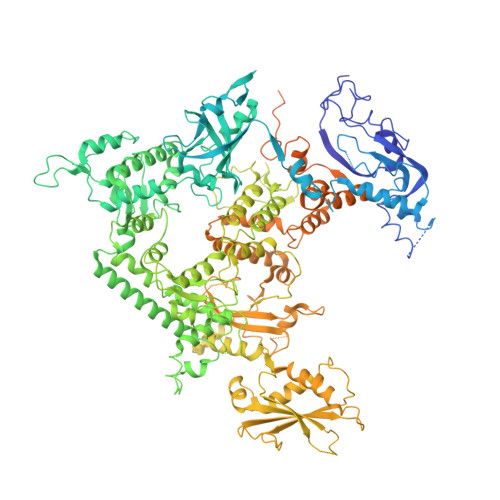
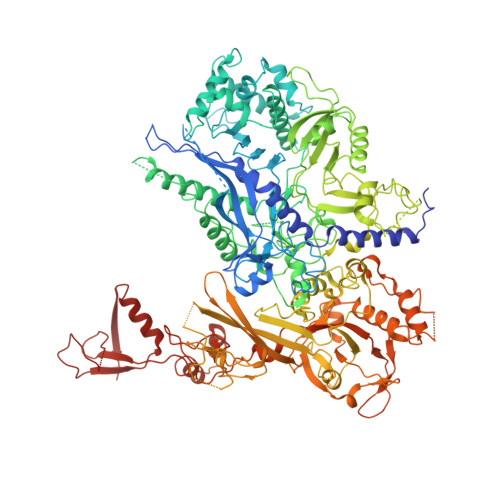
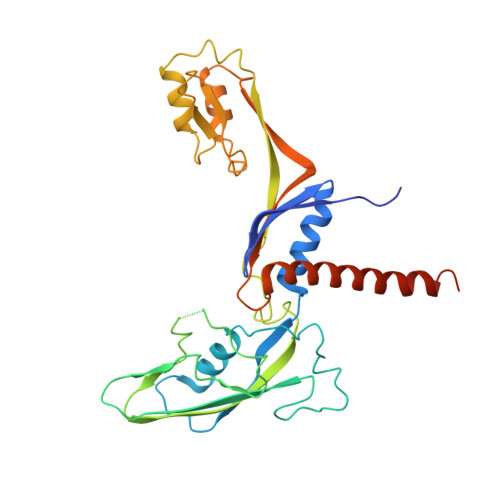
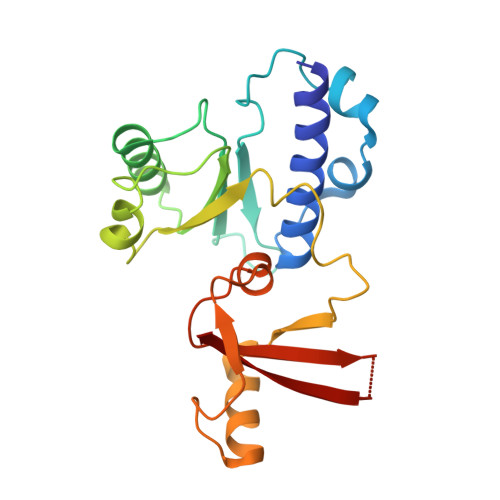
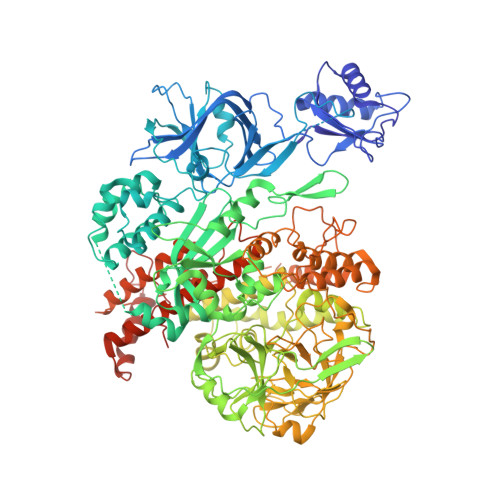
Pol IV and RDR2: A two-RNA-polymerase machine that produces double-stranded RNA.
Huang, K., Wu, X.X., Fang, C.L., Xu, Z.G., Zhang, H.W., Gao, J., Zhou, C.M., You, L.L., Gu, Z.X., Mu, W.H., Feng, Y., Wang, J.W., Zhang, Y.(2021) Science 374: 1579-1586
- PubMed: 34941388
- DOI: https://doi.org/10.1126/science.abj9184
- Primary Citation of Related Structures:
7EU0, 7EU1 - PubMed Abstract:
DNA methylation affects gene expression and maintains genome integrity. The DNA-dependent RNA polymerase IV (Pol IV), together with the RNA-dependent RNA polymerase RDR2, produces double-stranded small interfering RNA precursors essential for establishing and maintaining DNA methylation in plants. We determined the cryo–electron microscopy structures of the Pol IV–RDR2 holoenzyme and the backtracked transcription elongation complex. These structures reveal that Pol IV and RDR2 form a complex with their active sites connected by an interpolymerase channel, through which the Pol IV–generated transcript is handed over to the RDR2 active site after being backtracked, where it is used as the template for double-stranded RNA (dsRNA) synthesis. Our results describe a ‘backtracking-triggered RNA channeling’ mechanism underlying dsRNA synthesis and also shed light on the evolutionary trajectory of eukaryotic RNA polymerases.
- Key Laboratory of Synthetic Biology, CAS Center for Excellence in Molecular Plant Sciences, Shanghai Institute of Plant Physiology and Ecology, Chinese Academy of Sciences, Shanghai 200032, China.
Organizational Affiliation: