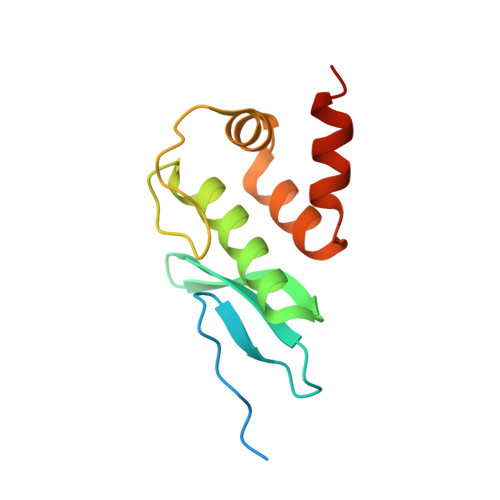
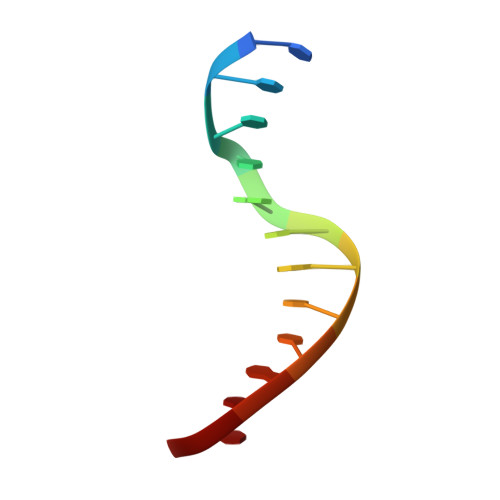
TEM1 combinatorially binds to FLOWERING LOCUS T and recruits a Polycomb factor to repress the floral transition in Arabidopsis.
Hu, H., Tian, S., Xie, G., Liu, R., Wang, N., Li, S., He, Y., Du, J.(2021) Proc Natl Acad Sci U S A 118
- PubMed: 34446554
- DOI: https://doi.org/10.1073/pnas.2103895118
- Primary Citation of Related Structures:
7ET4, 7ET5, 7ET6 - PubMed Abstract:
Arabidopsis TEMPRANILLO 1 (TEM1) is a transcriptional repressor that participates in multiple flowering pathways and negatively regulates the juvenile-to-adult transition and the flowering transition. To understand the molecular basis for the site-specific regulation of FLOWERING LOCUS T ( FT ) by TEM1, we determined the structures of the two plant-specific DNA-binding domains in TEM1, AP2 and B3, in complex with their target DNA sequences from the FT gene 5'-untranslated region (5'-UTR), revealing the molecular basis for TEM1 specificity for its DNA targets. In vitro binding assays revealed that the combination of the AP2 and B3 binding sites greatly enhanced the overall binding of TEM1 to the FT 5'-UTR, indicating TEM1 combinatorically recognizes the FT gene 5'-UTR. We further showed that TEM1 recruits the Polycomb repressive complex 2 (PRC2) to the FT 5'-UTR. The simultaneous binding of the TEM1 AP2 and B3 domains to FT is necessary for deposition of H3K27me3 at the FT 5'-UTR and for the flowering repressor function of TEM1. Overall, our data suggest that the combinatorial recognition of FT 5'-UTR by TEM1 ensures H3K27me3 deposition to precisely regulate the floral transition.
- National Key Laboratory of Plant Molecular Genetics and Shanghai Center for Plant Stress Biology, Center for Excellence in Molecular Plant Sciences, Chinese Academy of Sciences, Shanghai 201602, China.
Organizational Affiliation: