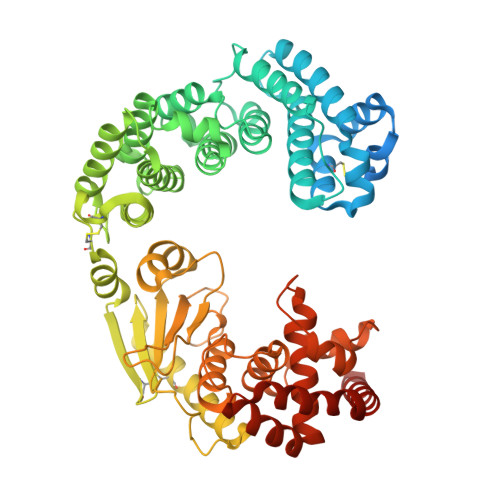
Structure of Vibrio collagenase VhaC provides insight into the mechanism of bacterial collagenolysis.
Wang, Y., Wang, P., Cao, H.Y., Ding, H.T., Su, H.N., Liu, S.C., Liu, G., Zhang, X., Li, C.Y., Peng, M., Li, F., Li, S., Chen, Y., Chen, X.L., Zhang, Y.Z.(2022) Nat Commun 13: 566-566
- PubMed: 35091565
- DOI: https://doi.org/10.1038/s41467-022-28264-1
- Primary Citation of Related Structures:
7ESI - PubMed Abstract:
The collagenases of Vibrio species, many of which are pathogens, have been regarded as an important virulence factor. However, there is little information on the structure and collagenolytic mechanism of Vibrio collagenase. Here, we report the crystal structure of the collagenase module (CM) of Vibrio collagenase VhaC and the conformation of VhaC in solution. Structural and biochemical analyses and molecular dynamics studies reveal that triple-helical collagen is initially recognized by the activator domain, followed by subsequent cleavage by the peptidase domain along with the closing movement of CM. This is different from the peptidolytic mode or the proposed collagenolysis of Clostridium collagenase. We propose a model for the integrated collagenolytic mechanism of VhaC, integrating the functions of VhaC accessory domains and its collagen degradation pattern. This study provides insight into the mechanism of bacterial collagenolysis and helps in structure-based drug design targeting of the Vibrio collagenase.
- State Key Laboratory of Microbial Technology, Shandong University, Qingdao, 266237, China.
Organizational Affiliation: