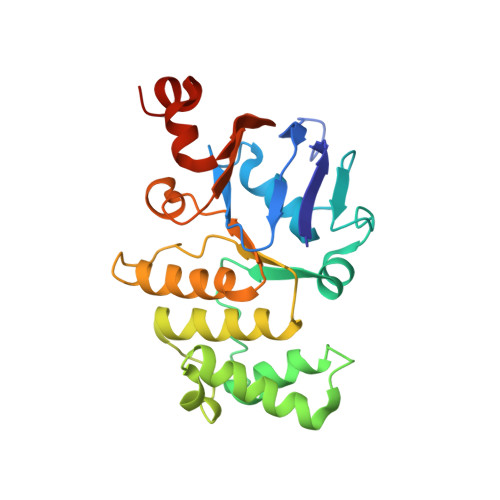
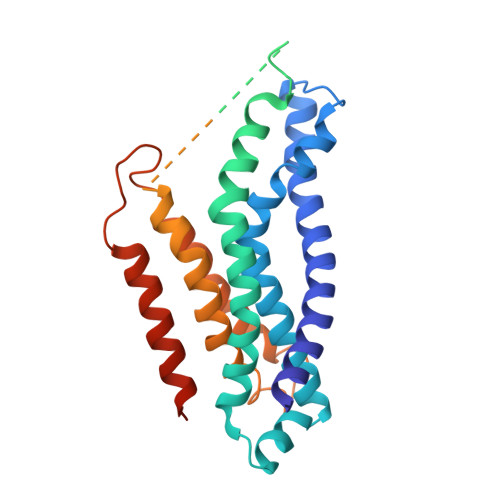
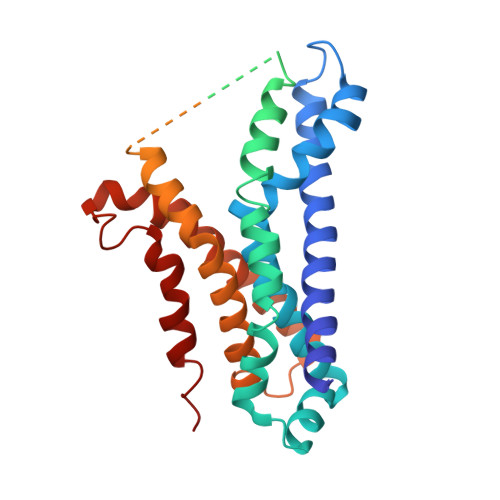
Cryo-EM structures of LptB 2 FG and LptB 2 FGC from Klebsiella pneumoniae in complex with lipopolysaccharide.
Luo, Q., Shi, H., Xu, X.(2021) Biochem Biophys Res Commun 571: 20-25
- PubMed: 34303191
- DOI: https://doi.org/10.1016/j.bbrc.2021.07.049
- Primary Citation of Related Structures:
7EFO - PubMed Abstract:
Lipopolysaccharide (LPS) is an essential component of the outer membrane (OM) in most Gram-negative bacteria. LPS transport from the inner membrane (IM) to the OM is achieved by seven lipopolysaccharide transport proteins (LptA-G). LptB 2 FG, an type VI ATP-binding cassette (ABC) transporter, forms a stable complex with LptC, extracts LPS from the IM and powers LPS transport to the OM. Here we report the cryo-EM structures of LptB 2 FG and LptB 2 FGC from Klebsiella pneumoniae in complex with LPS. The KpLptB 2 FG-LPS structure provides detailed interactions between LPS and the transporter, while the KpLptB 2 FGC-LPS structure may represent an intermediate state that the transmembrane helix of LptC has not been fully inserted into the transmembrane domains of LptB 2 FG.
- Department of Clinical Laboratory, Shenzhen Baoan Women's and Children's Hospital, Jinan University, Shenzhen, 518133, China; National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing, 100101, China. Electronic address: luoqingshan@moon.ibp.ac.cn.
Organizational Affiliation: