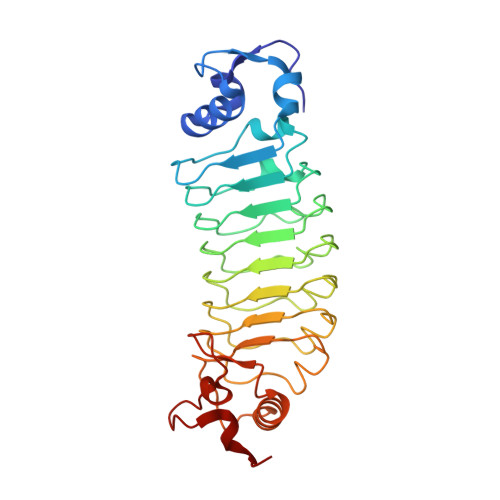
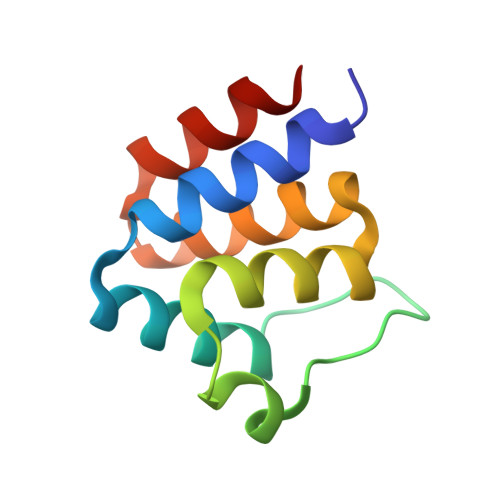
Oligomeric states of ASC specks regulate inflammatory responses by inflammasome in the extracellular space.
Yu, T.G., Cha, J.S., Kim, G., Sohn, Y.K., Yoo, Y., Kim, U., Song, J.J., Cho, H.S., Kim, H.S.(2023) Cell Death Discov 9: 142-142
- PubMed: 37120628
- DOI: https://doi.org/10.1038/s41420-023-01438-6
- Primary Citation of Related Structures:
7E5B - PubMed Abstract:
Inflammasomes are multi-protein complexes and play a crucial role in host defense against pathogens. Downstream inflammatory responses through inflammasomes are known to be related to the oligomerization degree of ASC specks, but the detailed mechanism still remains unexplored. Here, we demonstrate that oligomerization degrees of ASC specks regulate the caspase-1 activation in the extracellular space. A protein binder specific for a pyrin domain (PYD) of ASC (ASC PYD ) was developed, and structural analysis revealed that the protein binder effectively inhibits the interaction between PYDs, disassembling ASC specks into low oligomeric states. ASC specks with a low oligomerization degree were shown to enhance the activation of caspase-1 by recruiting and processing more premature caspase-1 through interactions between CARD of caspase-1 (caspase-1 CARD ) and CARD of ASC (ASC CARD ). These findings can provide insight into controlling the inflammasome-mediated inflammatory process as well as the development of inflammasome-targeting drugs.
- Departement of Biological Sciences, Korea Advanced Institute of Science and Technology (KAIST), Daejeon, 34141, Korea.
Organizational Affiliation: