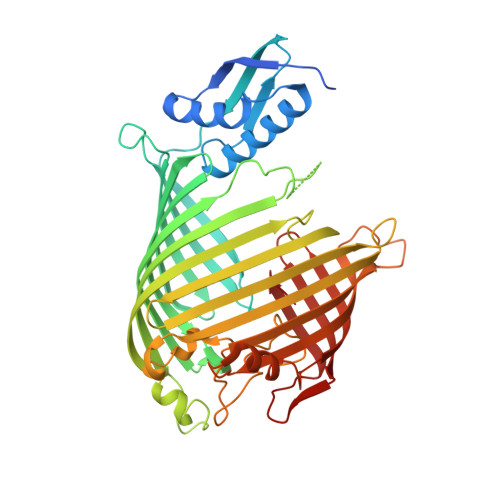
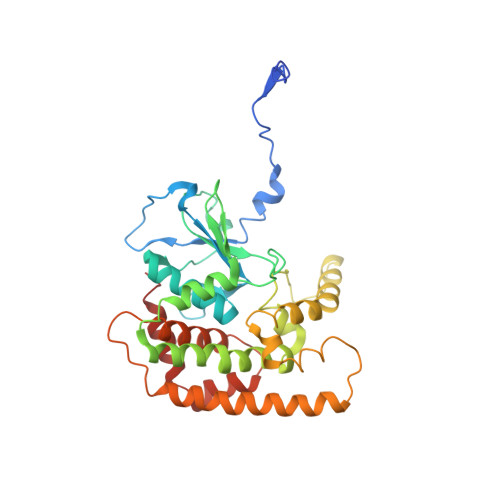
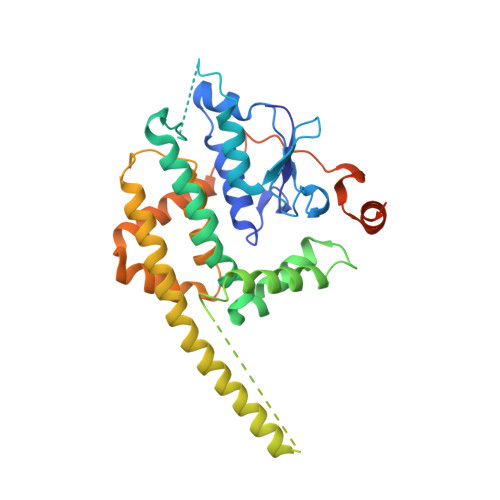
Structural insight into the SAM-mediated assembly of the mitochondrial TOM core complex.
Wang, Q., Guan, Z., Qi, L., Zhuang, J., Wang, C., Hong, S., Yan, L., Wu, Y., Cao, X., Cao, J., Yan, J., Zou, T., Liu, Z., Zhang, D., Yan, C., Yin, P.(2021) Science 373: 1377-1381
- PubMed: 34446444
- DOI: https://doi.org/10.1126/science.abh0704
- Primary Citation of Related Structures:
7E4H, 7E4I - PubMed Abstract:
β barrel outer membrane proteins (β-OMPs) play vital roles in mitochondria, chloroplasts, and Gram-negative bacteria. Evolutionarily conserved complexes such as the mitochondrial sorting and assembly machinery (SAM) mediate the assembly of β-OMPs. We investigated the SAM-mediated assembly of the translocase of the outer membrane (TOM) core complex. Cryo–electron microscopy structures of SAM–fully folded Tom40 and the SAM-Tom40/Tom5/Tom6 complexes at ~3-angstrom resolution reveal that Sam37 stabilizes the mature Tom40 mainly through electrostatic interactions, thus facilitating subsequent TOM assembly. These results support the β barrel switching model and provide structural insights into the assembly and release of β barrel complexes.
- National Key Laboratory of Crop Genetic Improvement, Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan 430070, China.
Organizational Affiliation: