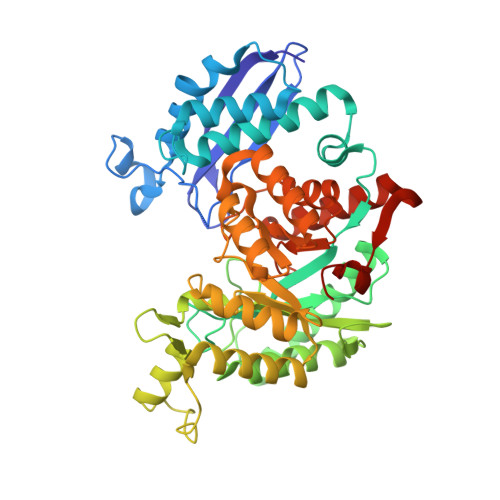
Evidence for the Rapid and Divergent Evolution of Mycoplasmas: Structural and Phylogenetic Analysis of Enolases.
Chen, R., Zhao, L., Gan, R., Feng, Z., Cui, C., Xie, X., Hao, F., Zhang, Z., Wang, L., Ran, T., Wang, W., Zhang, S., Li, Y., Zhang, W., Pang, M., Xiong, Q., Shao, G.(2022) Front Mol Biosci 8: 811106-811106