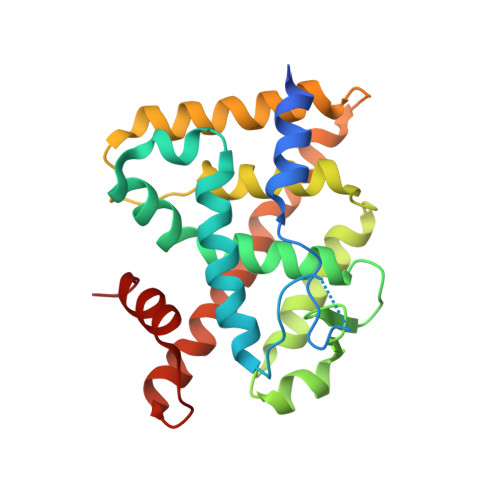
Discovery of a Novel Class of ERR alpha Agonists.
Shinozuka, T., Ito, S., Kimura, T., Izumi, M., Wakabayashi, K.(2021) ACS Med Chem Lett 12: 817-821
- PubMed: 34055231
- DOI: https://doi.org/10.1021/acsmedchemlett.1c00100
- Primary Citation of Related Structures:
7E2E - PubMed Abstract:
A novel class of estrogen-related receptor α (ERRα) agonists has been discovered. A structure-activity relationship study of high-throughput screening hits 1 and 2 led to the discovery of benzimidazole 3d (DS20362725) and acetophenone analogue 5c (DS45500853). The X-ray crystal structure of the ERRα ligand-binding domain in complex with 5c and PGC-1α coactivator peptide revealed conformational changes in the ligand-binding pocket to accommodate 5c and the key interaction between the protein and ligand. Since both analogues avoided PPARγ transcriptional activity, they can be useful tool compounds for investigating biological ERRα functions.
- R&D Division, Daiichi Sankyo Co., Ltd., 1-2-58 Hiromachi, Shinagawa-ku, Tokyo 140-8710, Japan.
Organizational Affiliation: