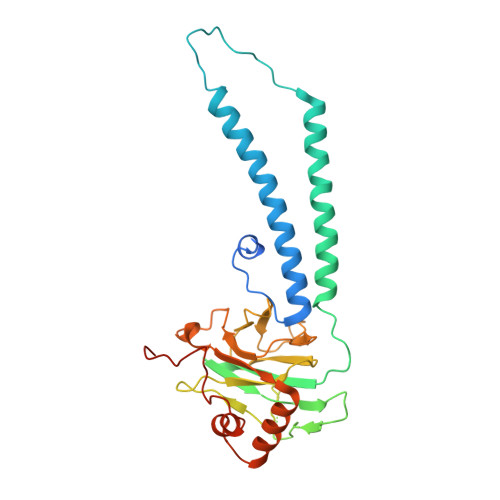
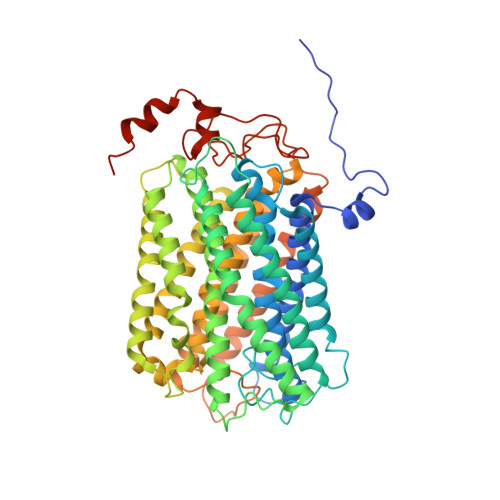
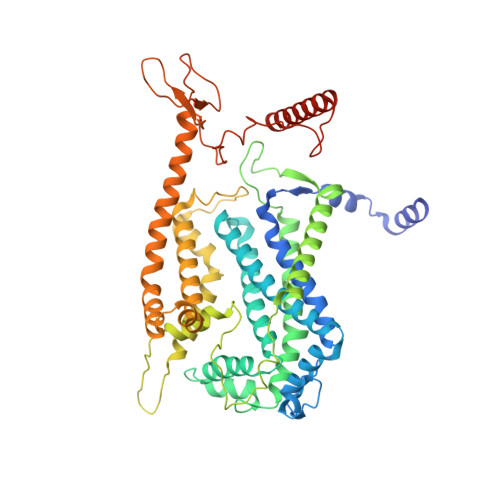
Structure of Mycobacterium tuberculosis cytochrome bcc in complex with Q203 and TB47, two anti-TB drug candidates.
Zhou, S., Wang, W., Zhou, X., Zhang, Y., Lai, Y., Tang, Y., Xu, J., Li, D., Lin, J., Yang, X., Ran, T., Chen, H., Guddat, L.W., Wang, Q., Gao, Y., Rao, Z., Gong, H.(2021) Elife 10
- PubMed: 34819223
- DOI: https://doi.org/10.7554/eLife.69418
- Primary Citation of Related Structures:
7E1V, 7E1W, 7E1X - PubMed Abstract:
Pathogenic mycobacteria pose a sustained threat to global human health. Recently, cytochrome bcc complexes have gained interest as targets for antibiotic drug development. However, there is currently no structural information for the cytochrome bcc complex from these pathogenic mycobacteria. Here, we report the structures of Mycobacterium tuberculosis cytochrome bcc alone (2.68 Å resolution) and in complex with clinical drug candidates Q203 (2.67 Å resolution) and TB47 (2.93 Å resolution) determined by single-particle cryo-electron microscopy. M. tuberculosis cytochrome bcc forms a dimeric assembly with endogenous menaquinone/menaquinol bound at the quinone/quinol-binding pockets. We observe Q203 and TB47 bound at the quinol-binding site and stabilized by hydrogen bonds with the side chains of QcrB Thr 313 and QcrB Glu 314 , residues that are conserved across pathogenic mycobacteria. These high-resolution images provide a basis for the design of new mycobacterial cytochrome bcc inhibitors that could be developed into broad-spectrum drugs to treat mycobacterial infections.
- State Key Laboratory of Medicinal Chemical Biology, College of Pharmacy, Nankai University, Tianjin, China.
Organizational Affiliation: