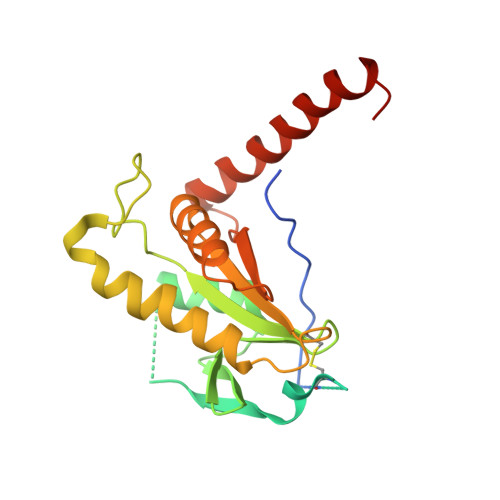
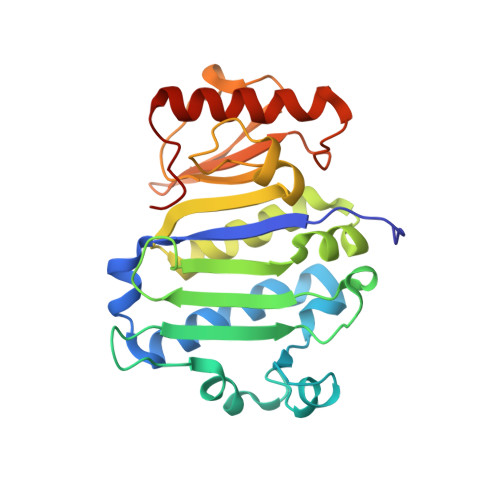
Structure and SAXS studies unveiled a novel inhibition mechanism of the Pseudomonas aeruginosa T6SS TseT-TsiT complex.
Wen, H., Liu, G., Geng, Z., Zhang, H., Li, Y., She, Z., Dong, Y.(2021) Int J Biol Macromol 188: 450-459
- PubMed: 34371041
- DOI: https://doi.org/10.1016/j.ijbiomac.2021.08.029
- Primary Citation of Related Structures:
7DYM - PubMed Abstract:
The bacterial type VI secretion system (T6SS) is a powerful arsenal that fires many toxic effectors into neighboring cells to gain advantage over inter-bacterial competition and eukaryotic host infection. Meanwhile, the cognate immunity proteins of these effectors are employed to protect themselves from the virulence. TseT-TsiT is a newly discovered effector-immunity (E-I) protein pair secreted by T6SS of Pseudomonas aeruginosa. Our group had reported the crystal structure of TsiT before. Here, we report the crystal structure of P. aeruginosa TseT-TsiT complex at 3.1 Å resolution. The interface of TseT-TsiT is characterized in this work. Through structure and small angle X-ray scattering (SAXS) studies, we discover that the long C-terminal helix of TseT may be flexible. Combining the homolog comparison results, we propose that TseT may form an oligomer in favor of its putative nuclease activity. Although TsiT doesn't directly block the putative active-site of TseT, it may hinder the TseT's oligomerization process to neutralize its virulence.
- Key Laboratory of Structural Biology, School of Life Sciences, University of Science and Technology of China, Hefei, China.
Organizational Affiliation: