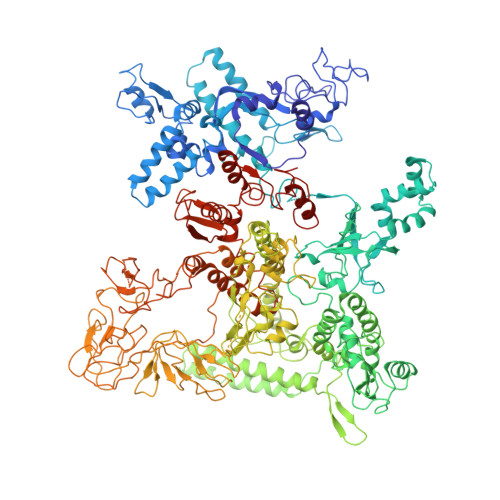
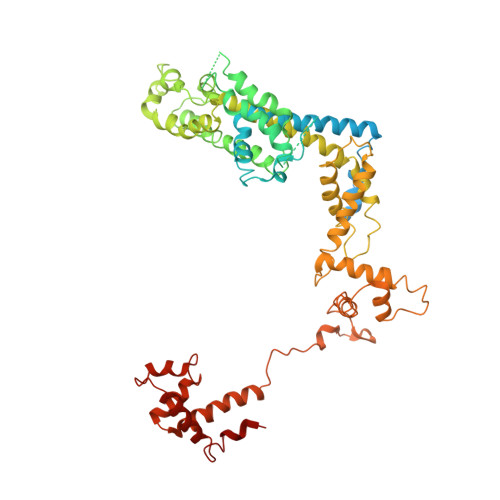
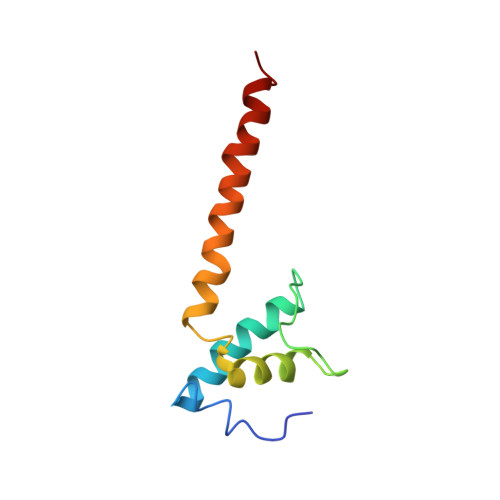
A unique binding between SspA and RNAP beta' NTH across low-GC Gram-negative bacteria facilitates SspA-mediated transcription regulation.
Wang, F., Feng, Y., Shang, Z., Lin, W.(2021) Biochem Biophys Res Commun 583: 86-92
- PubMed: 34735884
- DOI: https://doi.org/10.1016/j.bbrc.2021.10.048
- Primary Citation of Related Structures:
7DY6 - PubMed Abstract:
Stringent starvation protein A (SspA) involved in nucleotide metabolism, acid tolerance and virulence of bacteria has been demonstrated to function as a transcription factor to regulate σ 70 -dependent gene transcription through interacting with σ 70 region 4 and the zinc binding domain (ZBD) of E. coli RNA polymerase (EcoRNAP) β' subunit simultaneously. Despite extensive biochemical and structural analyses were reported recently, the interactions of SspA with RNAP are not comprehensively understood. Here, we reprocessed our previous cryo-EM dataset of EcoRNAP-promoter open complex with SspA (SspA-RPo) and obtained a significantly improved density map. Unexpectedly, the new map showed that SspA interacts with both N-terminal helix of β' subunit (β'ΝΤΗ) and ω subunit, which contributes to stabilize the SspA-EcoRNAP σ 70 holoenzyme complex. Sequence alignments and phylogenetic tree analyses of N-terminal sequences of β' subunit from different classes of bacteria revealed that β'ΝΤΗ is highly conserved and exclusively found in low-GC-content Gram-negative bacteria that harbor SspA, implying a co-evolution of β'ΝΤΗ and SspA. The transcription assays of wild-type SspA and its mutants demonstrated the interaction between SspA and β'ΝΤΗ facilitates the transcription regulation of SspA. Together, our results provide a more comprehensive insight into the interactions between SspA and RNAP and their roles in bacterial transcription regulation.
- Department of Pathogen Biology, School of Medicine & Holistic Integrative Medicine, Nanjing University of Chinese Medicine, Nanjing, China; State Key Laboratory of Natural Medicines, China Pharmaceutical University, Nanjing, China; Jiangsu Collaborative Innovation Center of Chinese Medicinal Resources Industrialization, Nanjing, 210023, China.
Organizational Affiliation: