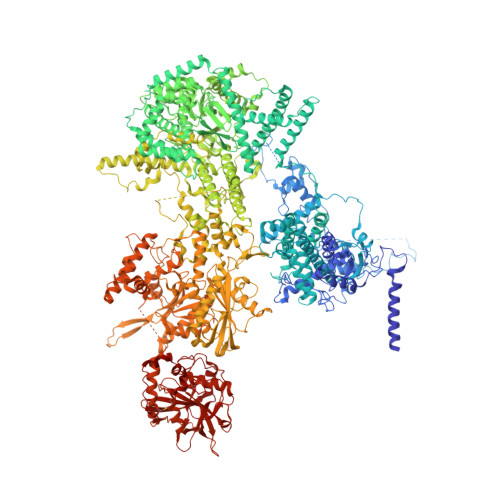
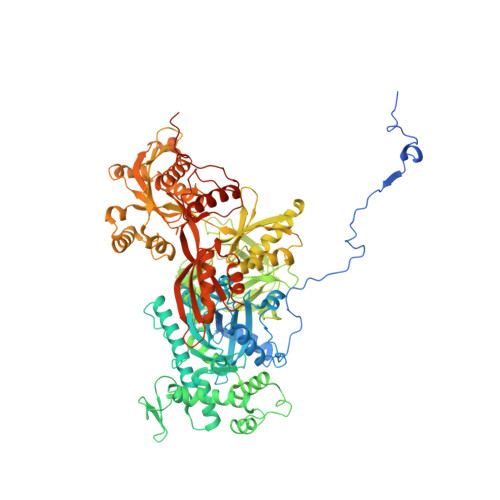
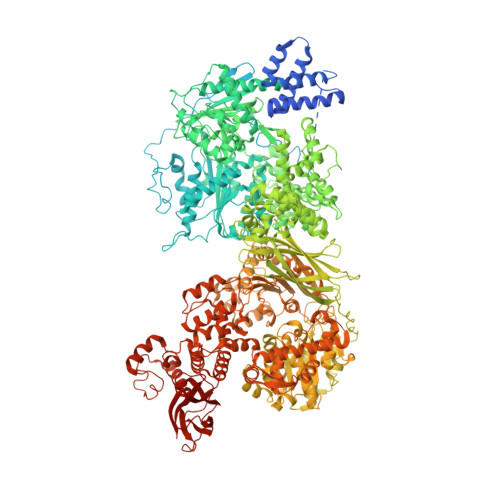
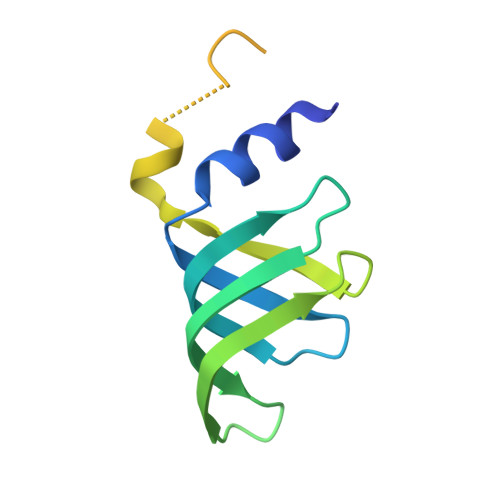
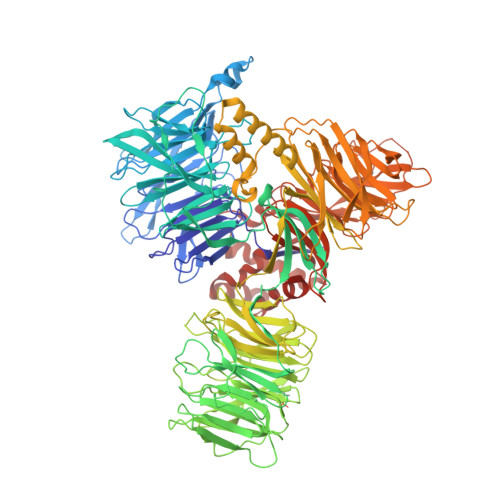
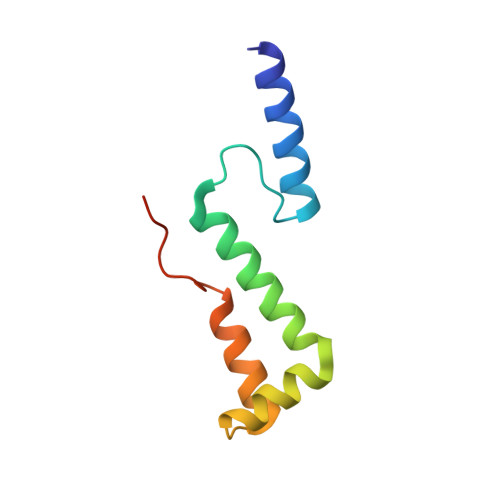
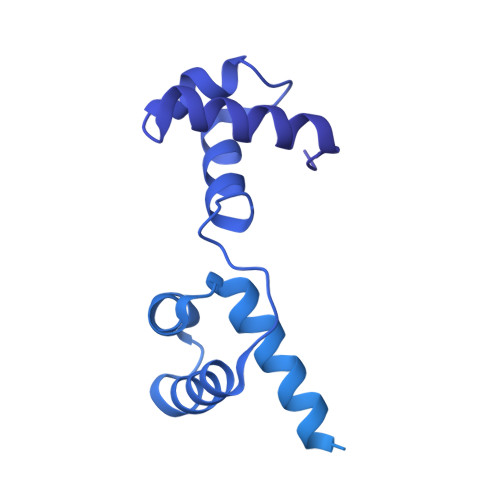
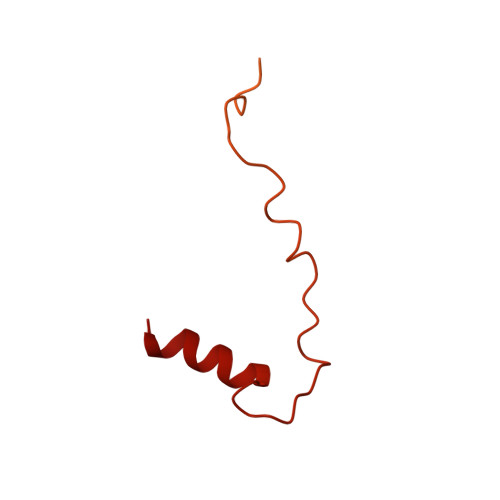
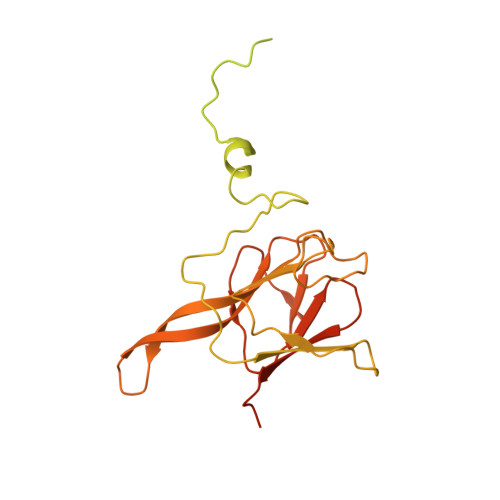
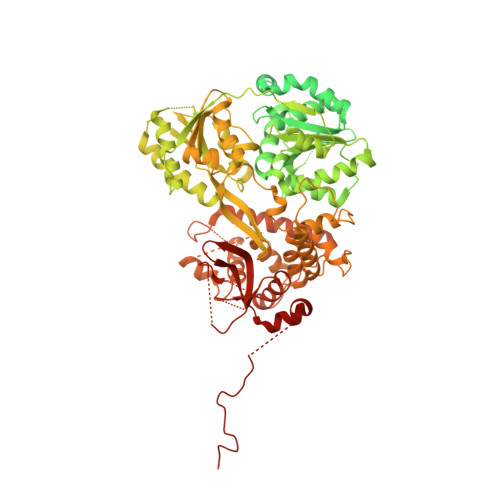
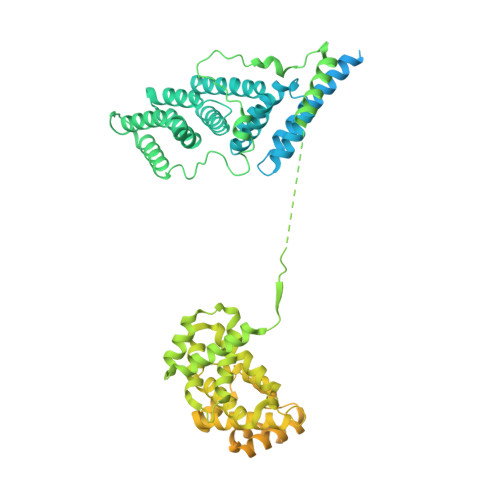
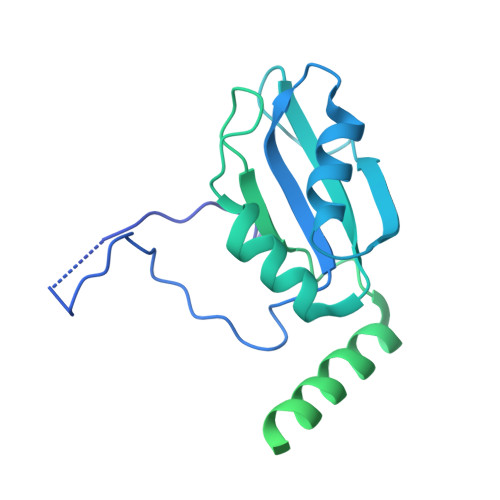
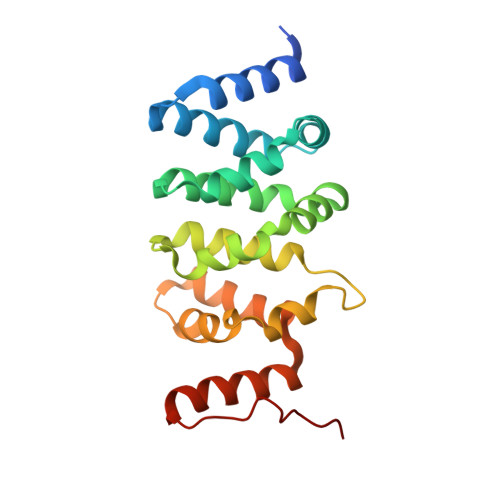
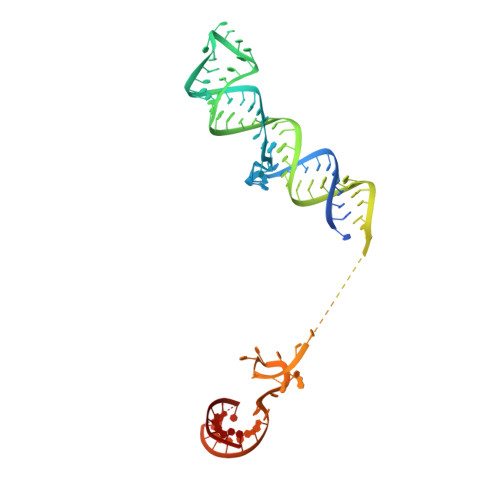
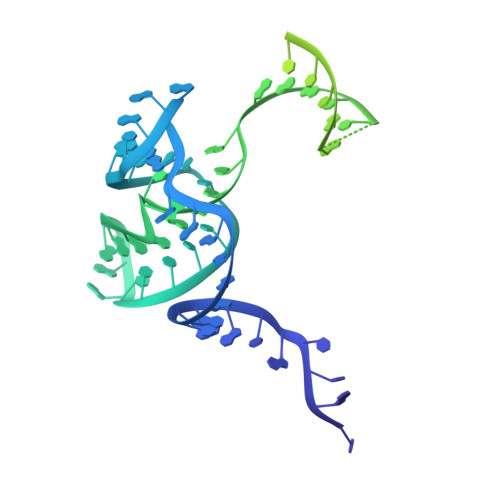
Structure of the activated human minor spliceosome.
Bai, R., Wan, R., Wang, L., Xu, K., Zhang, Q., Lei, J., Shi, Y.(2021) Science 371
- PubMed: 33509932
- DOI: https://doi.org/10.1126/science.abg0879
- Primary Citation of Related Structures:
7DVQ - PubMed Abstract:
The minor spliceosome mediates splicing of the rare but essential U12-type precursor messenger RNA. Here, we report the atomic features of the activated human minor spliceosome determined by cryo-electron microscopy at 2.9-angstrom resolution. The 5' splice site and branch point sequence of the U12-type intron are recognized by the U6atac and U12 small nuclear RNAs (snRNAs), respectively. Five newly identified proteins stabilize the conformation of the catalytic center: The zinc finger protein SCNM1 functionally mimics the SF3a complex of the major spliceosome, the RBM48-ARMC7 complex binds the γ-monomethyl phosphate cap at the 5' end of U6atac snRNA, the U-box protein PPIL2 coordinates loop I of U5 snRNA and stabilizes U5 small nuclear ribonucleoprotein (snRNP), and CRIPT stabilizes U12 snRNP. Our study provides a framework for the mechanistic understanding of the function of the human minor spliceosome.
- Key Laboratory of Structural Biology of Zhejiang Province, School of Life Sciences, Westlake University, Xihu District, Hangzhou 310024, Zhejiang Province, China.
Organizational Affiliation: