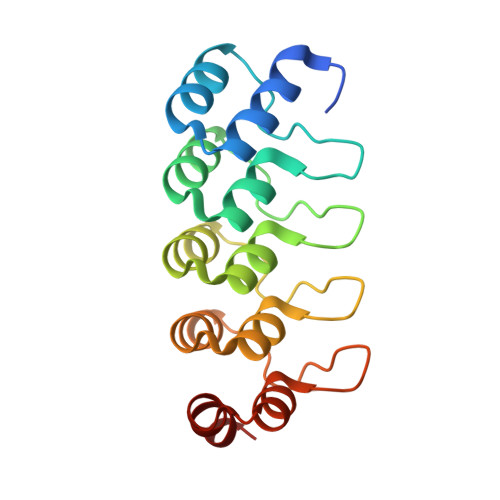
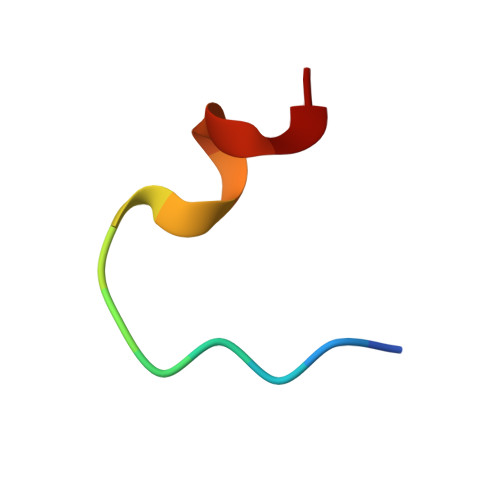
Distinct conformations of the HIV-1 V3 loop crown are targetable for broad neutralization.
Friedrich, N., Stiegeler, E., Glogl, M., Lemmin, T., Hansen, S., Kadelka, C., Wu, Y., Ernst, P., Maliqi, L., Foulkes, C., Morin, M., Eroglu, M., Liechti, T., Ivan, B., Reinberg, T., Schaefer, J.V., Karakus, U., Ursprung, S., Mann, A., Rusert, P., Kouyos, R.D., Robinson, J.A., Gunthard, H.F., Pluckthun, A., Trkola, A.(2021) Nat Commun 12: 6705-6705
- PubMed: 34795280
- DOI: https://doi.org/10.1038/s41467-021-27075-0
- Primary Citation of Related Structures:
7B4T, 7B4U, 7B4V, 7B4W, 7DNE, 7DNF, 7DNG - PubMed Abstract:
The V3 loop of the HIV-1 envelope (Env) protein elicits a vigorous, but largely non-neutralizing antibody response directed to the V3-crown, whereas rare broadly neutralizing antibodies (bnAbs) target the V3-base. Challenging this view, we present V3-crown directed broadly neutralizing Designed Ankyrin Repeat Proteins (bnDs) matching the breadth of V3-base bnAbs. While most bnAbs target prefusion Env, V3-crown bnDs bind open Env conformations triggered by CD4 engagement. BnDs achieve breadth by focusing on highly conserved residues that are accessible in two distinct V3 conformations, one of which resembles CCR5-bound V3. We further show that these V3-crown conformations can, in principle, be attacked by antibodies. Supporting this conclusion, analysis of antibody binding activity in the Swiss 4.5 K HIV-1 cohort (n = 4,281) revealed a co-evolution of V3-crown reactivities and neutralization breadth. Our results indicate a role of V3-crown responses and its conformational preferences in bnAb development to be considered in preventive and therapeutic approaches.
- Institute of Medical Virology, University of Zurich (UZH), Zurich, Switzerland.
Organizational Affiliation: