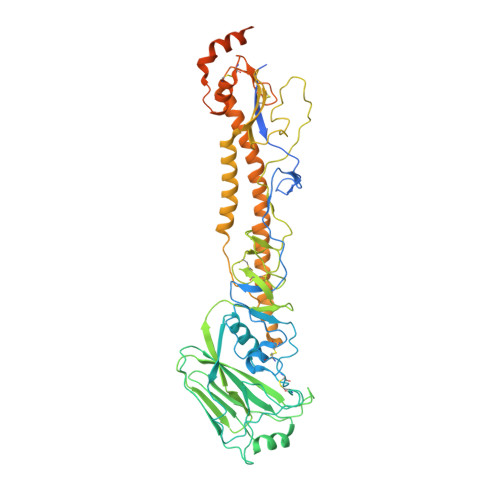
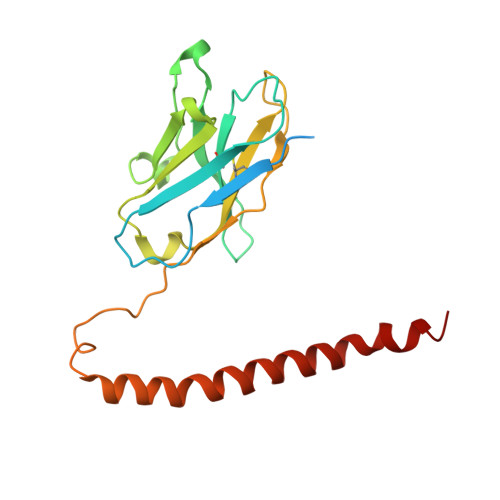
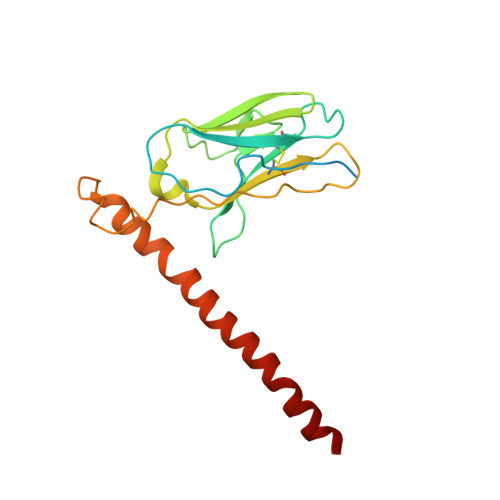
Hemagglutinin Influenza A virus (A/Okuda/1957(H2N2) bound with a neutralizing antibody
Makino-Okamura, C., Hata, H., Watanabe, T., Kim, S., Hebrard, M., Sato, R., Yan, M., Onodera, T., Takahashi, Y., Li, Q., Taylor, T., Okuno, Y., Kurosaki, T., Tomabechi, Y., Uchikubo-Kamo, T., Kamada, K., Shirouzu, M., Fukuyama, H.To be published.