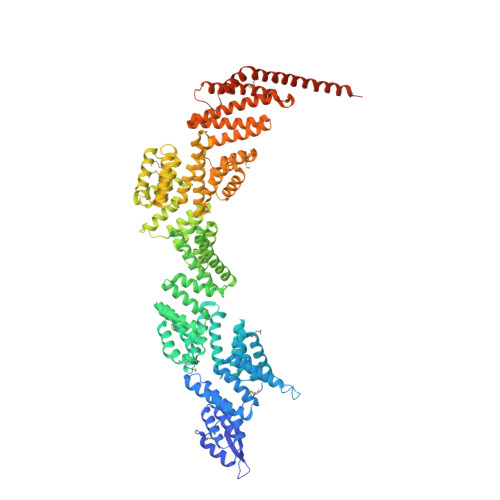
Crystal Structure of the Core Module of the Yeast Paf1 Complex.
Chen, F.L., Liu, B.B., Zeng, J.W., Guo, L., Ge, X., Feng, W., Li, D.F., Zhou, H., Long, J.F.(2021) J Mol Biology 434: 167369-167369
- PubMed: 34852272
- DOI: https://doi.org/10.1016/j.jmb.2021.167369
- Primary Citation of Related Structures:
7DKH - PubMed Abstract:
The highly conserved multifunctional polymerase-associated factor 1 (Paf1) complex (PAF1C), which consists of five core subunits: Ctr9, Paf1, Leo1, Cdc73, and Rtf1, acts as a diverse hub that regulates all stages of RNA polymerase II-mediated transcription and various other cellular functions. However, the underlying mechanisms remain unclear. Here, we report the crystal structure of the core module derived from a quaternary Ctr9/Paf1/Cdc73/Rtf1 complex of S. cerevisiae PAF1C, which reveals interfaces between the tetratricopeptide repeat module in Ctr9 and Cdc73 or Rtf1, and find that the Ctr9/Paf1 subcomplex is the key scaffold for PAF1C assembly. Our study demonstrates that Cdc73 binds Ctr9/Paf1 subcomplex with a very similar conformation within thermophilic fungi or human PAF1C, and that the binding of Cdc73 to PAF1C is important for yeast growth. Importantly, our structure reveals for the first time that the extreme C-terminus of Rtf1 adopts an "L"-shaped structure, which interacts with Ctr9 specifically. In addition, disruption of the binding of either Cdc73 or Rtf1 to PAF1C greatly affects the normal level of histone H2B K123 monoubiquitination in vivo. Collectively, our results provide a structural insight into the architecture of the quaternary Ctr9/Paf1/Cdc73/Rtf1 complex and PAF1C functional regulation.
- State Key Laboratory of Medicinal Chemical Biology, Tianjin Key Laboratory of Protein Science, and College of Life Sciences, Nankai University, 94 Weijin Road, Tianjin 300071, China.
Organizational Affiliation: