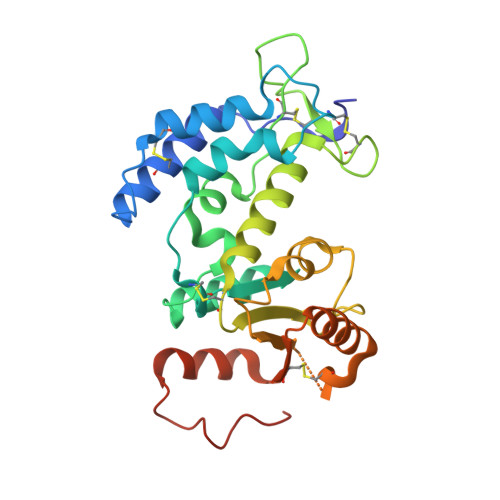
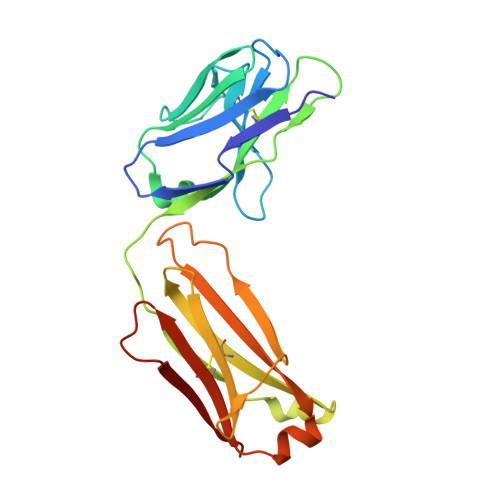
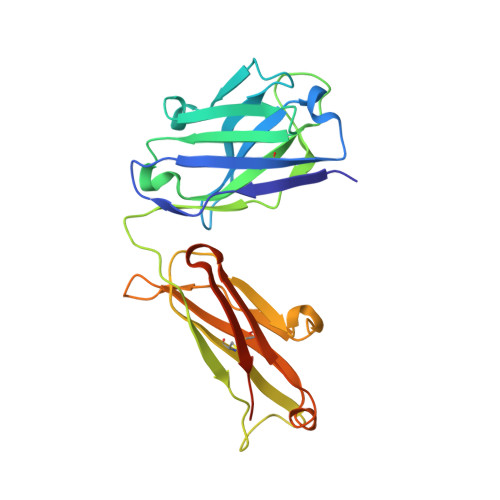
Crystal structure of CD38 in complex with daratumumab, a first-in-class anti-CD38 antibody drug for treating multiple myeloma.
Lee, H.T., Kim, Y., Park, U.B., Jeong, T.J., Lee, S.H., Heo, Y.S.(2021) Biochem Biophys Res Commun 536: 26-31
- PubMed: 33360095
- DOI: https://doi.org/10.1016/j.bbrc.2020.12.048
- Primary Citation of Related Structures:
7DHA - PubMed Abstract:
Multiple myeloma is a blood cancer characterized by the plasma cell malignancy in the bone marrow, resulting in the destruction of bone tissue. Recently, the US FDA approved two antibody drugs for the treatment of multiple myeloma, daratumumab and isatuximab, targeting CD38, a type II transmembrane glycoprotein highly expressed in plasma cells and multiple myeloma cells. Here, we report the crystal structure of CD38 in complex with the Fab fragment of daratumumab, providing its exact epitope on CD38 and the structural insights into the mechanism of action of the antibody drug. Daratumumab binds to a specific discontinuous region on CD38 that includes residues located opposite to the active site of CD38. All the six complementarity determining regions of daratumumab are involved in the CD38 interaction. The epitopes of daratumumab and isatuximab do not overlap at all and their bindings to CD38 induce different structural changes within the CD38 protein. This structural study can facilitate the design of improved biologics or effective combination therapies for the treatment of multiple myeloma.
- Department of Chemistry, Konkuk University, 120 Neungdong-ro, Gwangjin-gu, Seoul, 05029, Republic of Korea.
Organizational Affiliation: