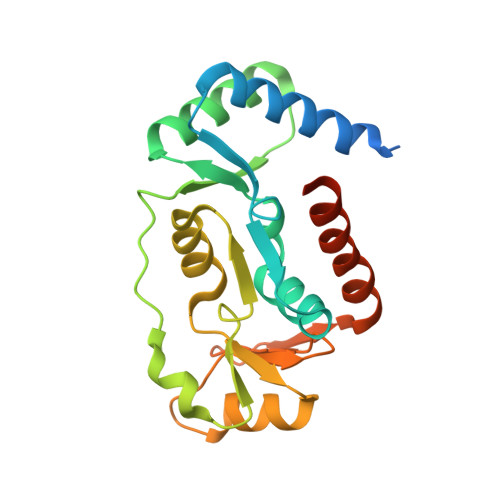
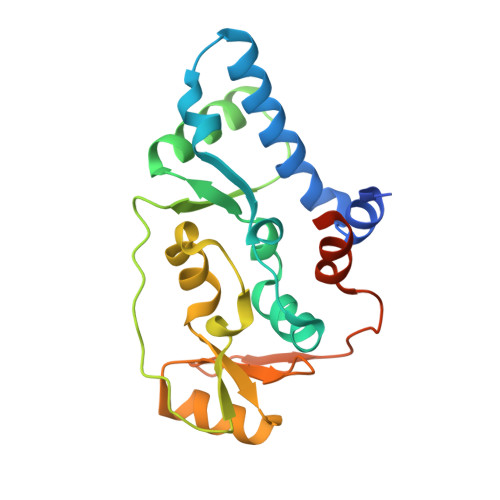
Folding of cohesin's coiled coil is important for Scc2/4-induced association with chromosomes.
Petela, N.J., Gonzalez Llamazares, A., Dixon, S., Hu, B., Lee, B.G., Metson, J., Seo, H., Ferrer-Harding, A., Voulgaris, M., Gligoris, T., Collier, J., Oh, B.H., Lowe, J., Nasmyth, K.A.(2021) Elife 10
- PubMed: 34259632
- DOI: https://doi.org/10.7554/eLife.67268
- Primary Citation of Related Structures:
7DG5, 7OGT - PubMed Abstract:
Cohesin's association with and translocation along chromosomal DNAs depend on an ATP hydrolysis cycle driving the association and subsequent release of DNA. This involves DNA being 'clamped' by Scc2 and ATP-dependent engagement of cohesin's Smc1 and Smc3 head domains. Scc2's replacement by Pds5 abrogates cohesin's ATPase and has an important role in halting DNA loop extrusion. The ATPase domains of all SMC proteins are separated from their hinge dimerisation domains by 50-nm-long coiled coils, which have been observed to zip up along their entire length and fold around an elbow, thereby greatly shortening the distance between hinges and ATPase heads. Whether folding exists in vivo or has any physiological importance is not known. We present here a cryo-EM structure of the apo form of cohesin that reveals the structure of folded and zipped-up coils in unprecedented detail and shows that Scc2 can associate with Smc1's ATPase head even when it is fully disengaged from that of Smc3. Using cysteine-specific crosslinking, we show that cohesin's coiled coils are frequently folded in vivo, including when cohesin holds sister chromatids together. Moreover, we describe a mutation ( SMC1D588Y ) within Smc1's hinge that alters how Scc2 and Pds5 interact with Smc1's hinge and that enables Scc2 to support loading in the absence of its normal partner Scc4. The mutant phenotype of loading without Scc4 is only explicable if loading depends on an association between Scc2/4 and cohesin's hinge, which in turn requires coiled coil folding.
- Department of Biochemistry, University of Oxford, Oxford, United Kingdom.
Organizational Affiliation: