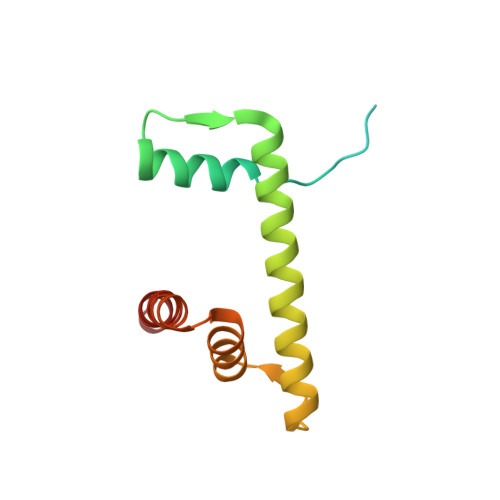
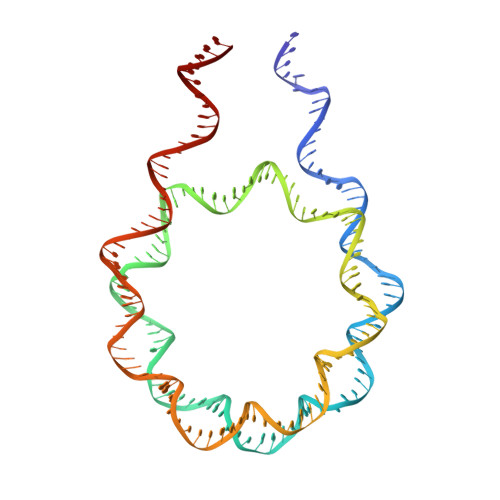
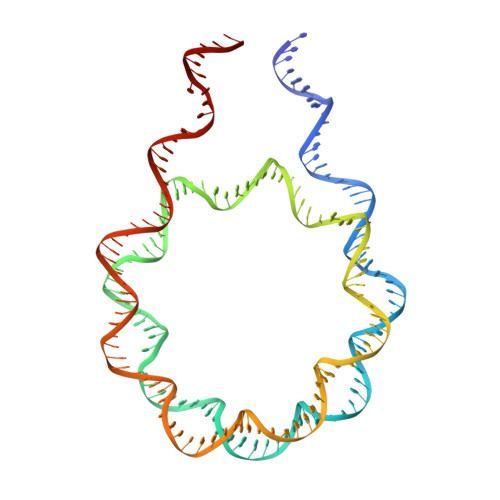
Linker histone defines structure and self-association behaviour of the 177 bp human chromatosome.
Wang, S., Vogirala, V.K., Soman, A., Berezhnoy, N.V., Liu, Z.B., Wong, A.S.W., Korolev, N., Su, C.J., Sandin, S., Nordenskiold, L.(2021) Sci Rep 11: 380-380
- PubMed: 33432055
- DOI: https://doi.org/10.1038/s41598-020-79654-8
- Primary Citation of Related Structures:
7DBP - PubMed Abstract:
Linker histones play essential roles in the regulation and maintenance of the dynamic chromatin structure of higher eukaryotes. The influence of human histone H1.0 on the nucleosome structure and biophysical properties of the resulting chromatosome were investigated and compared with the 177-bp nucleosome using Cryo-EM and SAXS. The 4.5 Å Cryo-EM chromatosome structure showed that the linker histone binds at the nucleosome dyad interacting with both linker DNA arms but in a tilted manner leaning towards one of the linker sides. The chromatosome is laterally compacted and rigid in the dyad and linker DNA area, in comparison with the nucleosome where linker DNA region is more flexible and displays structural variability. In solution, the chromatosomes appear slightly larger than the nucleosomes, with the volume increase compared to the bound linker histone, according to solution SAXS measurements. SAXS X-ray diffraction characterisation of Mg-precipitated samples showed that the different shapes of the 177 chromatosome enabled the formation of a highly ordered lamello-columnar phase when precipitated by Mg 2+ , indicating the influence of linker histone on the nucleosome stacking. The biological significance of linker histone, therefore, may be affected by the change in the polyelectrolyte and DNA conformation properties of the chromatosomes, in comparison to nucleosomes.
- School of Biological Sciences, Nanyang Technological University, 60 Nanyang Dr., Singapore, 637551, Singapore.
Organizational Affiliation: