Engineering Disulphide-Free Autonomous Antibody VH Domains to modulate intracellular pathways
Frosi, Y., Lin, Y.C., Jiang, S., Ramlan, S.R., Hew, K., Asial, I., Engman, A.H., Goh, M., Thean, D., Nordlund, P., Lane, D.P., Brown, C.J.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Eukaryotic translation initiation factor 4E | 217 | Homo sapiens | Mutation(s): 0 Gene Names: EIF4E, EIF4EL1, EIF4F | 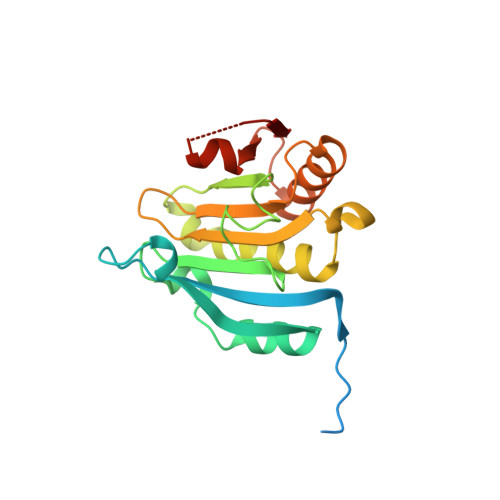 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P06730 (Homo sapiens) Explore P06730 Go to UniProtKB: P06730 | |||||
PHAROS: P06730 GTEx: ENSG00000151247 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P06730 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| VH Domain (1C5) | 161 | Homo sapiens | Mutation(s): 0 | 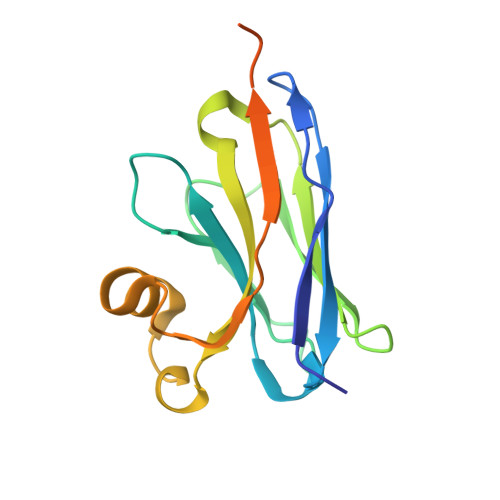 | |
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| MGO Query on MGO | C [auth A] | [[(2R,3S,4R,5R)-5-(6-AMINO-3-METHYL-4-OXO-5H-IMIDAZO[4,5-C]PYRIDIN-1-YL)-3,4-DIHYDROXY-OXOLAN-2-YL]METHOXY-HYDROXY-PHOSPHORYL] PHOSPHONO HYDROGEN PHOSPHATE C12 H20 N4 O14 P3 LPHBJZOLBGBNJF-XYHAGOFUSA-O |  | ||
| MES Query on MES | D [auth A] | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID C6 H13 N O4 S SXGZJKUKBWWHRA-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 198.848 | α = 90 |
| b = 45.324 | β = 93.192 |
| c = 39.763 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| XDS | data reduction |
| XDS | data scaling |
| PHASER | phasing |