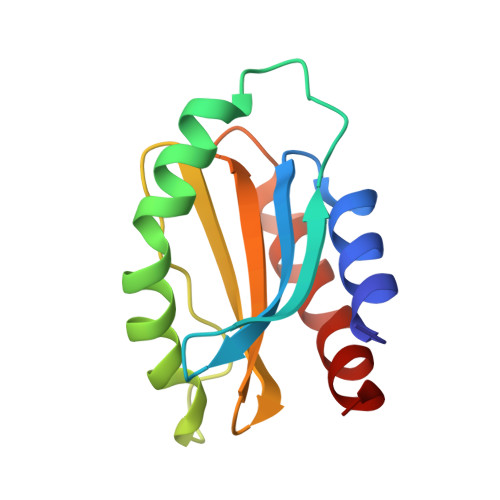
Structural characterization of Myxococcus xanthus MglC, a component of the polarity control system, and its interactions with its paralog MglB.
Kapoor, S., Kodesia, A., Kalidas, N., Thakur, K.G.(2021) J Biological Chem : 100308-100308
- PubMed: 33493516
- DOI: https://doi.org/10.1016/j.jbc.2021.100308
- Primary Citation of Related Structures:
7CT3, 7CY1 - PubMed Abstract:
The δ-proteobacteria Myxococcus xanthus displays social (S) and adventurous (A) motilities, which require pole-to-pole reversal of the motility regulator proteins. Mutual gliding motility protein C (MglC), a paralog of GTPase-activating protein Mutual gliding motility protein B (MglB), is a member of the polarity module involved in regulating motility. However, little is known about the structure and function of MglC. Here, we determined ∼1.85 Å resolution crystal structure of MglC using Selenomethionine Single-wavelength anomalous diffraction. The crystal structure revealed that, despite sharing <9% sequence identity, both MglB and MglC adopt a Regulatory Light Chain 7 family fold. However, MglC has a distinct ∼30° to 40° shift in the orientation of the functionally important α2 helix compared with other structural homologs. Using isothermal titration calorimetry and size-exclusion chromatography, we show that MglC binds MglB in 2:4 stoichiometry with submicromolar range dissociation constant. Using small-angle X-ray scattering and molecular docking studies, we show that the MglBC complex consists of a MglC homodimer sandwiched between two homodimers of MglB. A combination of size-exclusion chromatography and site-directed mutagenesis studies confirmed the MglBC interacting interface obtained by molecular docking studies. Finally, we show that the C-terminal region of MglB, crucial for binding its established partner MglA, is not required for binding MglC. These studies suggest that the MglB uses distinct interfaces to bind MglA and MglC. Based on these data, we propose a model suggesting a new role for MglC in polarity reversal in M. xanthus.
- Structural Biology Laboratory, Council of Scientific and Industrial Research-Institute of Microbial Technology, G. N. Ramachandran Protein Centre, Chandigarh, India.
Organizational Affiliation: