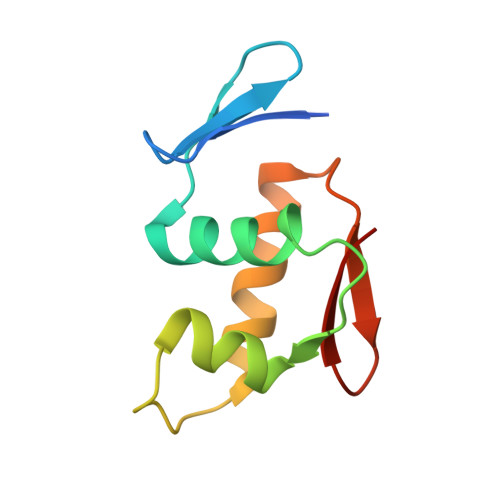
Crystal structure of the DNA-binding domain of Bacillus subtilis CssR.
Dahal, P., Kim, D.Y., Kwon, E.(2021) Biochem Biophys Res Commun 555: 26-31
- PubMed: 33812055
- DOI: https://doi.org/10.1016/j.bbrc.2021.03.101
- Primary Citation of Related Structures:
7CX5 - PubMed Abstract:
Bacteria utilize two-component systems to regulate gene expression in response to changes in environmental stimuli. CssS/CssR, a two-component system in Bacillus subtilis, is responsible for overcoming envelope stresses caused by heat shock and secretion overload. During stress, the sensor component CssS is auto-phosphorylated and transfers the phosphoryl group to the response regulator CssR. Phosphorylated CssR then directly regulates the transcription of genes required to counteract the stress. Here, the crystal structure of the DNA-binding domain of CssR, determined at 1.07 Å resolution, is reported. The structure shows that the DNA-binding domain of CssR harbors a winged helix-turn-helix motif that is conserved in the OmpR/PhoB subfamily of response regulators. Based on the crystal structure, the dimeric architecture of the full-length CssR and its DNA-binding mode were suggested.
- College of Pharmacy, Yeungnam University, Gyeongsan, Gyeongbuk, 38541, Republic of Korea.
Organizational Affiliation: