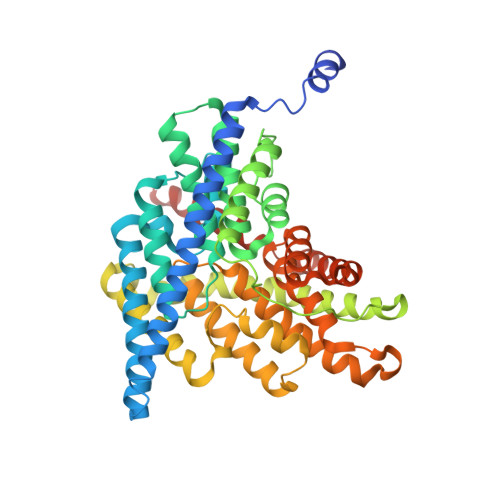
Altering CLC stoichiometry by reducing non-polar side-chains at the dimerization interface.
Mersch, K., Ozturk, T.N., Park, K., Lim, H.H., Robertson, J.L.(2021) J Mol Biology 433: 166886-166886
- PubMed: 33617898
- DOI: https://doi.org/10.1016/j.jmb.2021.166886
- Primary Citation of Related Structures:
7CVS, 7CVT - PubMed Abstract:
CLC-ec1 is a Cl - /H + antiporter that forms stable homodimers in lipid bilayers, with a free energy of -10.9 kcal/mol in 2:1 POPE/POPG lipid bilayers. The dimerization interface is formed by four transmembrane helices: H, I, P and Q, that are lined by non-polar side-chains that come in close contact, yet it is unclear as to whether their interactions drive dimerization. To investigate whether non-polar side-chains are required for dimer assembly, we designed a series of constructs where side-chain packing in the dimer state is significantly reduced by making 4-5 alanine substitutions along each helix (H-ala, I-ala, P-ala, Q-ala). All constructs are functional and three purify as stable dimers in detergent micelles despite the removal of significant side-chain interactions. On the other hand, H-ala shows the unique behavior of purifying as a mixture of monomers and dimers, followed by a rapid and complete conversion to monomers. In lipid bilayers, all four constructs are monomeric as examined by single-molecule photobleaching analysis. Further study of the H-helix shows that the single mutation L194A is sufficient to yield monomeric CLC-ec1 in detergent micelles and lipid bilayers. X-ray crystal structures of L194A reveal the protein re-assembles to form dimers, with a structure that is identical to wild-type. Altogether, these results demonstrate that non-polar membrane embedded side-chains play an important role in defining dimer stability, but the stoichiometry is highly contextual to the solvent environment. Furthermore, we discovered that L194 is a molecular hot-spot for defining dimerization of CLC-ec1.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine in St. Louis, St. Louis, MO, USA; Department of Molecular Physiology and Biophysics, The University of Iowa, Iowa City, IA, USA.
Organizational Affiliation: