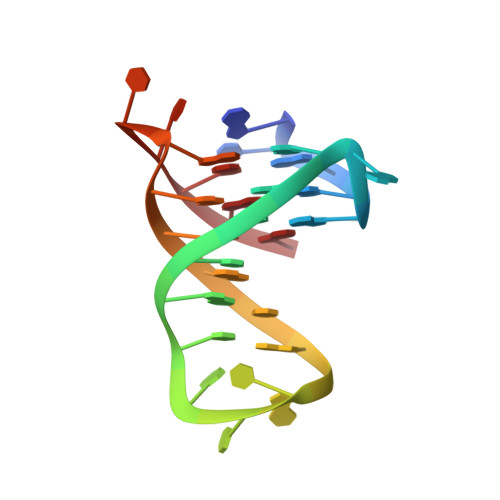
Coexistence of two quadruplex-duplex hybrids in the PIM1 gene.
Tan, D.J.Y., Winnerdy, F.R., Lim, K.W., Phan, A.T.(2020) Nucleic Acids Res 48: 11162-11171
- PubMed: 32976598
- DOI: https://doi.org/10.1093/nar/gkaa752
- Primary Citation of Related Structures:
7CV3, 7CV4 - PubMed Abstract:
The triple-negative breast cancer (TNBC), a subtype of breast cancer which lacks of targeted therapies, exhibits a poor prognosis. It was shown recently that the PIM1 oncogene is highly related to the proliferation of TNBC cells. A quadruplex-duplex hybrid (QDH) forming sequence was recently found to exist near the transcription start site of PIM1. This structure could be an attractive target for regulation of the PIM1 gene expression and thus the treatment of TNBC. Here, we present the solution structures of two QDHs that could coexist in the human PIM1 gene. Form 1 is a three-G-tetrad-layered (3+1) G-quadruplex containing a propeller loop, a lateral loop and a stem-loop made up of three G•C Watson-Crick base pairs. On the other hand, Form 2 is an anti-parallel G-quadruplex comprising two G-tetrads and a G•C•G•C tetrad; the structure has three lateral loops with the middle stem-loop made up of two Watson-Crick G•C base pairs. These structures provide valuable information for the design of G-quadruplex-specific ligands for PIM1 transcription regulation.
- School of Physical and Mathematical Sciences, Nanyang Technological University, Singapore 637371, Singapore.
Organizational Affiliation: