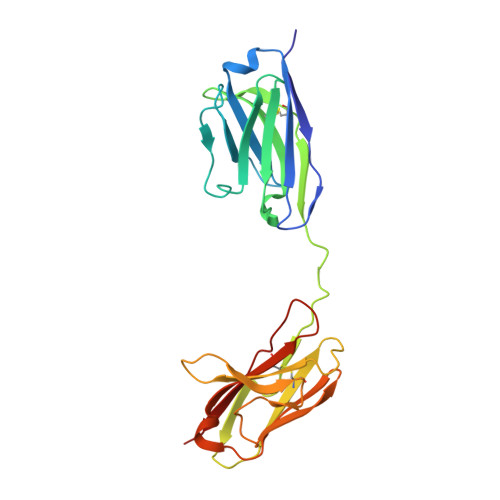
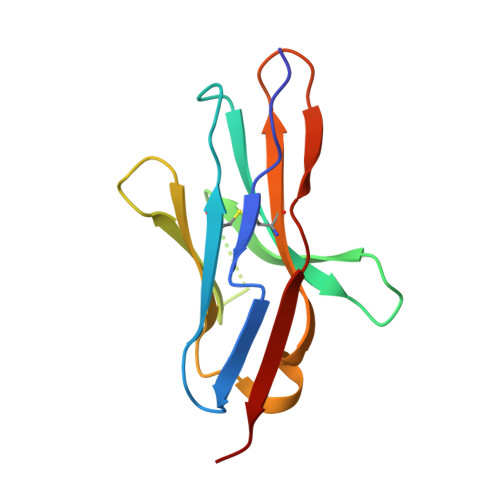
N-glycosylation of PD-1 promotes binding of camrelizumab.
Liu, K., Tan, S., Jin, W., Guan, J., Wang, Q., Sun, H., Qi, J., Yan, J., Chai, Y., Wang, Z., Deng, C., Gao, G.F.(2020) EMBO Rep 21: e51444-e51444
- PubMed: 33063473
- DOI: https://doi.org/10.15252/embr.202051444
- Primary Citation of Related Structures:
7CU5 - PubMed Abstract:
PD-1 is a highly glycosylated inhibitory receptor expressed mainly on T cells. Targeting of PD-1 with monoclonal antibodies (MAbs) to block the interaction with its ligand PD-L1 has been successful for the treatment of multiple tumors. However, polymorphisms at N-glycosylation sites of PD-1 exist in the human population that might affect antibody binding, and dysregulated glycosylation has been observed in the tumor microenvironment. Here, we demonstrate varied N-glycan composition in PD-1, and show that the binding affinity of camrelizumab, a recently approved PD-1-specific MAb, to non-glycosylated PD-1 proteins from E. coli is substantially decreased compared with glycosylated PD-1. The structure of the camrelizumab/PD-1 complex reveals that camrelizumab mainly utilizes its heavy chain to bind to PD-1, while the light chain sterically inhibits the binding of PD-L1 to PD-1. Glycosylation of asparagine 58 (N58) promotes the interaction with camrelizumab, while the efficiency of camrelizumab to inhibit the binding of PD-L1 is substantially reduced for glycosylation-deficient PD-1. These results increase our understanding of how glycosylation affects the activity of PD-1-specific MAbs during immune checkpoint therapy.
- Faculty of Health Sciences, University of Macau, Macau SAR, China.
Organizational Affiliation: