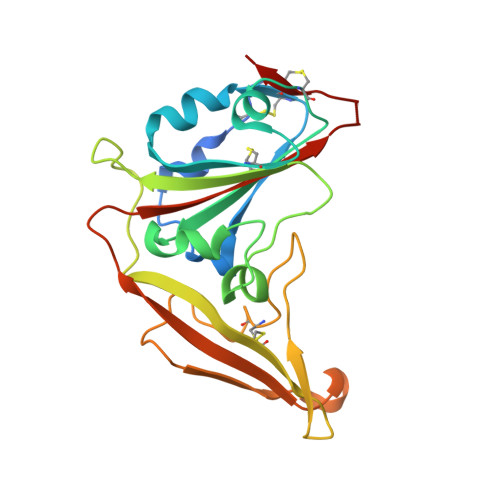
The structure of a novel antibody against the spike protein inhibits Middle East respiratory syndrome coronavirus infections.
Jang, T.H., Park, W.J., Lee, H., Woo, H.M., Lee, S.Y., Kim, K.C., Kim, S.S., Hong, E., Song, J., Lee, J.Y.(2022) Sci Rep 12: 1260-1260
- PubMed: 35075213
- DOI: https://doi.org/10.1038/s41598-022-05318-4
- Primary Citation of Related Structures:
7COE - PubMed Abstract:
Middle East respiratory syndrome coronavirus (MERS-CoV) is a zoonotic virus, responsible for outbreaks of a severe respiratory illness in humans with a fatality rate of 30%. Currently, there are no vaccines or United States food and drug administration (FDA)-approved therapeutics for humans. The spike protein displayed on the surface of MERS-CoV functions in the attachment and fusion of virions to host cellular membranes and is the target of the host antibody response. Here, we provide a molecular method for neutralizing MERS-CoV through potent antibody-mediated targeting of the receptor-binding subdomain (RBD) of the spike protein. The structural characterization of the neutralizing antibody (KNIH90-F1) complexed with RBD using X-ray crystallography revealed three critical epitopes (D509, R511, and E513) in the RBD region of the spike protein. Further investigation of MERS-CoV mutants that escaped neutralization by the antibody supported the identification of these epitopes in the RBD region. The neutralizing activity of this antibody is solely provided by these specific molecular structures. This work should contribute to the development of vaccines or therapeutic antibodies for MERS-CoV.
- New Drug Development Center, Daegu-Gyeongbuk Medical Innovation Foundation, Daegu, Republic of Korea.
Organizational Affiliation: