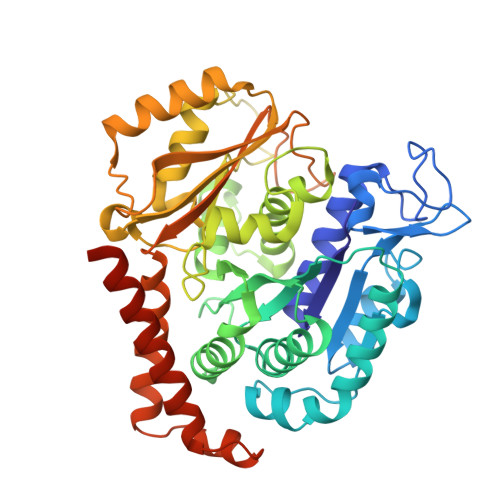
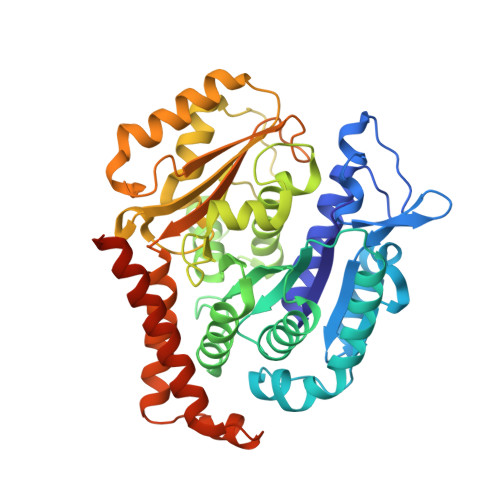
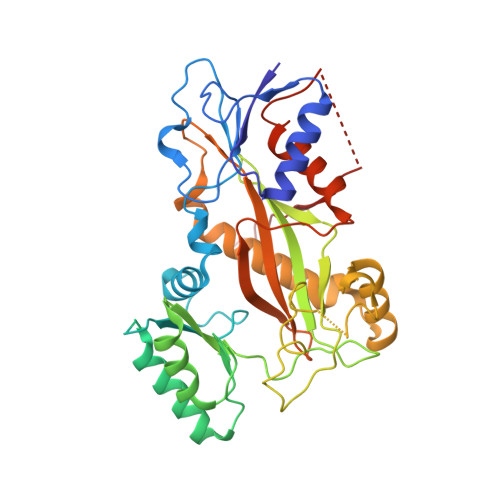
The high-resolution X-ray structure of vinca-domain inhibitors of microtubules provides a rational approach for drug design.
Chengyong, W., Jinghong, X., Yanyan, W., Qing-Jie, X., Lingling, M., Yuyan, L., Hai, C., Qian, L., Quan, Z., Bo, S., Yuxi, W.(2021) FEBS Lett 595: 195-205
- PubMed: 33220079
- DOI: https://doi.org/10.1002/1873-3468.14003
- Primary Citation of Related Structures:
7CNM, 7CNN, 7CNO - PubMed Abstract:
Tubulin vinca-domain ligands can inhibit microtubule polymerization, causing cell death in mitosis, and their potential against multiple cancer types has been demonstrated. However, due to drug resistance and toxicities, development of novel vinca-domain ligands is still needed. In this study, we determined the high-resolution crystal structures of vinorelbine, YXD, and Phomopsin A in complex with tubulin at 2.5 Å. Additionally, we recapitulated all previously published high-resolution crystal structures of the vinca binding site to reveal critical residues and the molecular mechanism of vinca-domain ligands interacting with tubulin. Furthermore, we designed putatively novel triazolopyrimidine derivatives by introducing secondary amine groups to establish salt-bridge and H-bond interactions with Asp179 β1 and Asn329 α2 . Our studies provided the structural basis for designing novel tubulin vinca-domain ligands.
- Cancer Center, West China Hospital, Sichuan University, Chengdu, China.
Organizational Affiliation: