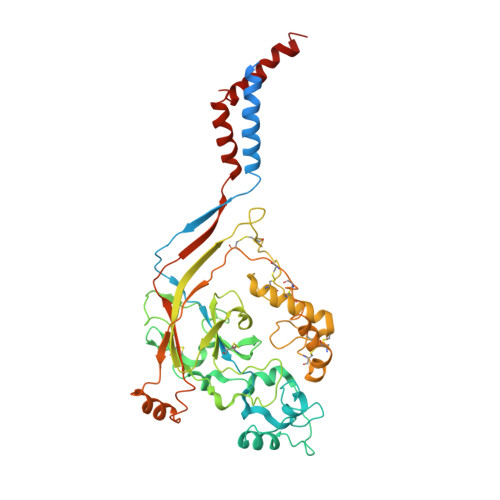
Structural insights into human acid-sensing ion channel 1a inhibition by snake toxin mambalgin1.
Sun, D., Liu, S., Li, S., Zhang, M., Yang, F., Wen, M., Shi, P., Wang, T., Pan, M., Chang, S., Zhang, X., Zhang, L., Tian, C., Liu, L.(2020) Elife 9
- PubMed: 32915133
- DOI: https://doi.org/10.7554/eLife.57096
- Primary Citation of Related Structures:
7CFS, 7CFT - PubMed Abstract:
Acid-sensing ion channels (ASICs) are proton-gated cation channels that are involved in diverse neuronal processes including pain sensing. The peptide toxin Mambalgin1 (Mamba1) from black mamba snake venom can reversibly inhibit the conductance of ASICs, causing an analgesic effect. However, the detailed mechanism by which Mamba1 inhibits ASIC1s, especially how Mamba1 binding to the extracellular domain affects the conformational changes of the transmembrane domain of ASICs remains elusive. Here, we present single-particle cryo-EM structures of human ASIC1a (hASIC1a) and the hASIC1a-Mamba1 complex at resolutions of 3.56 and 3.90 Å, respectively. The structures revealed the inhibited conformation of hASIC1a upon Mamba1 binding. The combination of the structural and physiological data indicates that Mamba1 preferentially binds hASIC1a in a closed state and reduces the proton sensitivity of the channel, representing a closed-state trapping mechanism.
- Hefei National Laboratory of Physical Sciences at Microscale, Anhui Laboratory of Advanced Photonic Science and Technology and School of Life Sciences, University of Science and Technology of China, Hefei, China.
Organizational Affiliation: