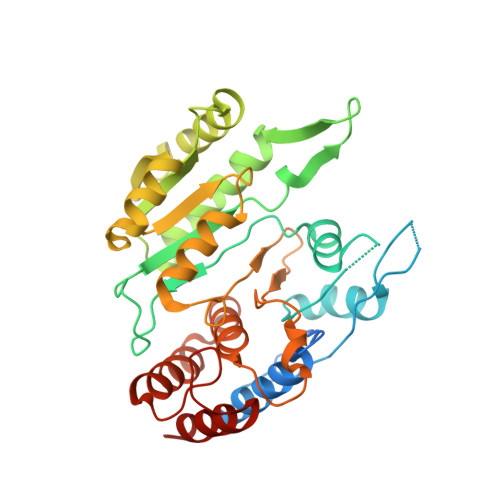
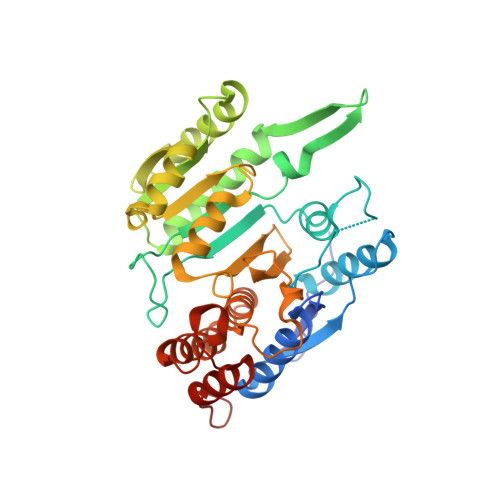
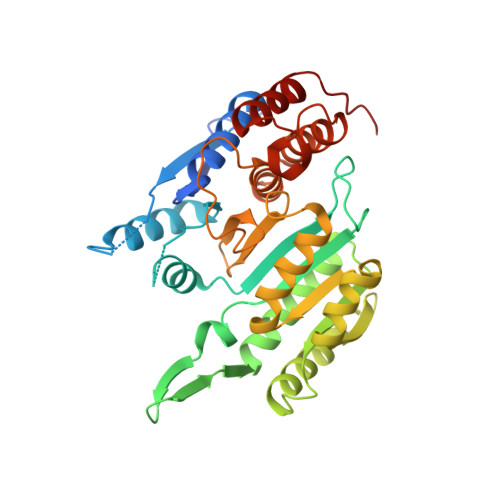
Structure and allosteric regulation of human NAD-dependent isocitrate dehydrogenase.
Sun, P., Liu, Y., Ma, T., Ding, J.(2020) Cell Discov 6: 94-94
- PubMed: 33349631
- DOI: https://doi.org/10.1038/s41421-020-00220-7
- Primary Citation of Related Structures:
7CE3 - PubMed Abstract:
Human NAD-dependent isocitrate dehydrogenase or HsIDH3 catalyzes the decarboxylation of isocitrate into α-ketoglutarate in the TCA cycle. HsIDH3 exists and functions as a heterooctamer composed of the αβ and αγ heterodimers, and is regulated allosterically and/or competitively by numerous metabolites including CIT, ADP, ATP, and NADH. In this work, we report the crystal structure of HsIDH3 containing a β mutant in apo form. In the HsIDH3 structure, the αβ and αγ heterodimers form the α 2 βγ heterotetramer via their clasp domains, and two α 2 βγ heterotetramers form the (α 2 βγ) 2 heterooctamer through insertion of the N-terminus of the γ subunit of one heterotetramer into the back cleft of the β subunit of the other heterotetramer. The functional roles of the key residues at the allosteric site, the pseudo allosteric site, the heterodimer and heterodimer-heterodimer interfaces, and the N-terminal of the γ subunit are validated by mutagenesis and kinetic studies. Our structural and biochemical data together demonstrate that the allosteric site plays an important role but the pseudo allosteric site plays no role in the allosteric activation of the enzyme; the activation signal from the allosteric site is transmitted to the active sites of both αβ and αγ heterodimers via the clasp domains; and the N-terminal of the γ subunit plays a critical role in the formation of the heterooctamer to ensure the optimal activity of the enzyme. These findings reveal the molecular mechanism of the assembly and allosteric regulation of HsIDH3.
- State Key Laboratory of Molecular Biology, Shanghai Institute of Biochemistry and Cell Biology, Center for Excellence in Molecular Cell Science, University of Chinese Academy of Sciences, Chinese Academy of Sciences, 320 Yueyang Road, Shanghai 200031, China.
Organizational Affiliation: