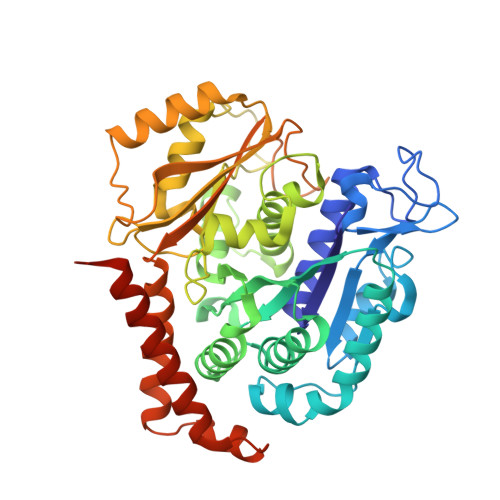
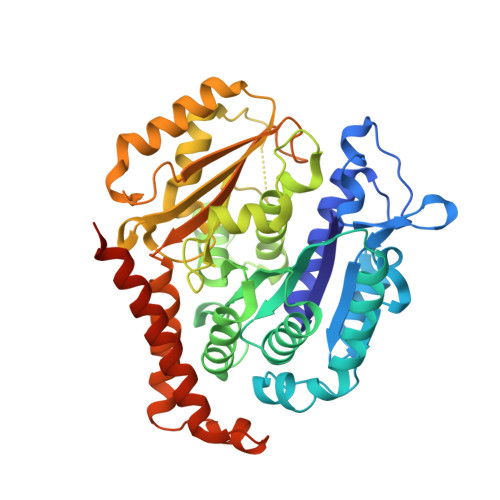
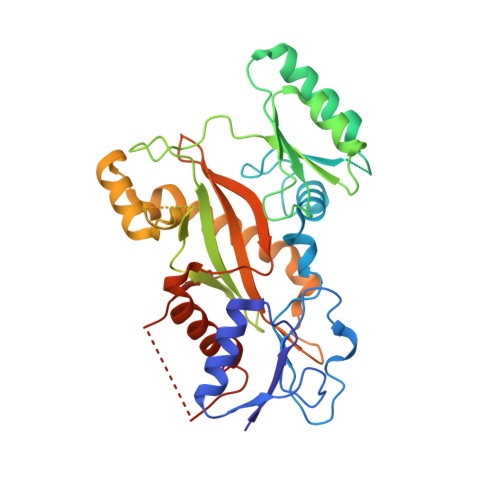
Design, Synthesis, and Bioactivity Evaluation of Dual-Target Inhibitors of Tubulin and Src Kinase Guided by Crystal Structure.
Wang, L., Zheng, Y., Li, D., Yang, J., Lei, L., Yan, W., Zheng, W., Tang, M., Shi, M., Zhang, R., Cai, X., Ni, H., Ma, X., Li, N., Hong, F., Ye, H., Chen, L.(2021) J Med Chem 64: 8127-8141
- PubMed: 34081857
- DOI: https://doi.org/10.1021/acs.jmedchem.0c01961
- Primary Citation of Related Structures:
7CBZ - PubMed Abstract:
Klisyri (KX01) is a dual tubulin/Src protein inhibitor that has shown potential therapeutic effects in several tumor models. However, a phase II clinical trial in patients with bone-metastatic castration-resistant prostate cancer was halted because of lack of efficacy. We previously reported that KX01 binds to the colchicine site of β-tubulin and its morpholine group lies close to α-tubulin's surface. Thus, we hypothesized that enhancing the interaction of KX01 with α-tubulin could increase tubulin inhibition and synthesized a series of KX01 derivatives directed by docking studies. Among these derivatives, 8a exhibited more than 10-fold antiproliferation activity in several tumor cells than KX01 and significantly improved in vivo antitumor effects. The X-ray crystal structure suggested that 8a both bound to the colchicine site and extended into the interior of α-tubulin to form potent interactions, presenting a novel binding mode. A potential clinical candidate for cancer therapy was identified in this study.
- State Key Laboratory of Biotherapy/Collaborative Innovation Center of Biotherapy and Cancer Center, West China Hospital of Sichuan University, Chengdu 610041, China.
Organizational Affiliation: