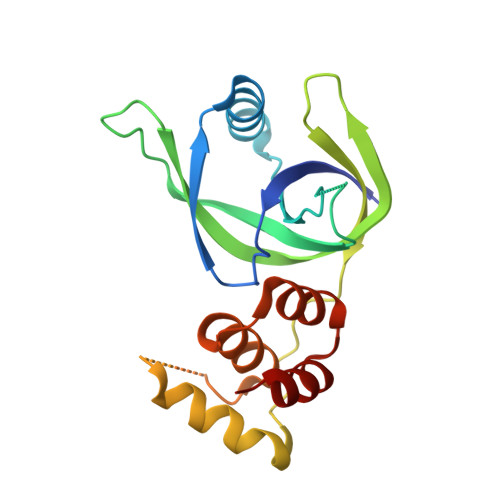
Crystal structure of XCC3289 from Xanthomonas campestris: homology with the N-terminal substrate-binding domain of Lon peptidase.
Singh, R., Deshmukh, S., Kumar, A., Goyal, V.D., Makde, R.D.(2020) Acta Crystallogr F Struct Biol Commun 76: 488-494
- PubMed: 33006577
- DOI: https://doi.org/10.1107/S2053230X20011875
- Primary Citation of Related Structures:
7CAY - PubMed Abstract:
LonA peptidase is a major component of the protein quality-control mechanism in both prokaryotes and the organelles of eukaryotes. Proteins homologous to the N-terminal domain of LonA peptidase, but lacking its other domains, are conserved in several phyla of prokaryotes, including the Xanthomonadales order. However, the function of these homologous proteins (LonNTD-like proteins) is not known. Here, the crystal structure of the LonNTD-like protein from Xanthomonas campestris (XCC3289; UniProt Q8P5P7) is reported at 2.8 Å resolution. The structure was solved by molecular replacement and contains one polypeptide in the asymmetric unit. The structure was refined to an R free of 29%. The structure of XCC3289 consists of two domains joined by a long loop. The N-terminal domain (residues 1-112) consists of an α-helix surrounded by β-sheets, whereas the C-terminal domain (residues 123-193) is an α-helical bundle. The fold and spatial orientation of the two domains closely resembles those of the N-terminal domains of the LonA peptidases from Escherichia coli and Mycobacterium avium. The structure is also similar to that of cereblon, a substrate-recognizing component of the E3 ubiquitin ligase complex. The N-terminal domains of both LonA and cereblon are known to be involved in specific protein-protein interactions. This structural analysis suggests that XCC3289 and other LonNTD-like proteins might also be capable of such protein-protein interactions.
- Beamline Development and Application Section, Bhabha Atomic Research Centre, Mumbai 400 085, India.
Organizational Affiliation: