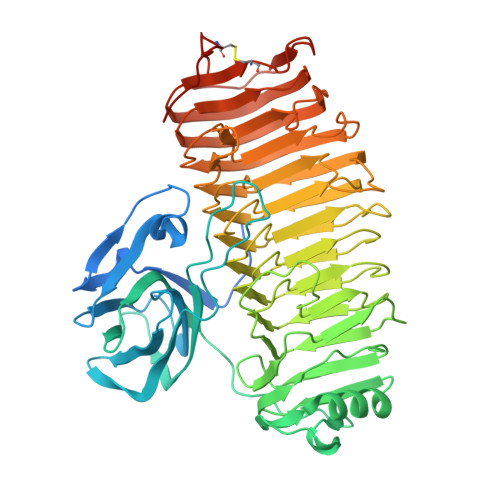
Crystal structure of the catalytic unit of thermostable GH87 alpha-1,3-glucanase from Streptomyces thermodiastaticus strain HF3-3.
Itoh, T., Panti, N., Hayashi, J., Toyotake, Y., Matsui, D., Yano, S., Wakayama, M., Hibi, T.(2020) Biochem Biophys Res Commun 533: 1170-1176
- PubMed: 33041007
- DOI: https://doi.org/10.1016/j.bbrc.2020.09.133
- Primary Citation of Related Structures:
7C7D - PubMed Abstract:
α-1,3-Glucan is a homopolymer composed of D-glucose (Glc) and it is an extracellular polysaccharide found in dental plaque due to Streptococcus species. α-1,3-Glucanase from Streptomyces thermodiastaticus strain HF3-3 (Agl-ST) has been identified as a thermostable α-1,3-glucanase, which is classified into glycoside hydrolase family 87 (GH87) and specifically hydrolyzes α-1,3-glucan with an endo-action. The enzyme has a potential to inhibit the production of dental plaque and to be used for biotechnological applications. Here we show the structure of the catalytic unit of Agl-ST determined at 1.16 Å resolution using X-ray crystallography. The catalytic unit is composed of two modules, a β-sandwich fold module, and a right-handed β-helix fold module, which resembles other structural characterized GH87 enzymes from Bacillus circulans str. KA-304 and Paenibacillus glycanilyticus str. FH11, with moderate sequence identities between each other (approximately 27% between the catalytic units). However, Agl-ST is smaller in size and more thermally stable than the others. A disulfide bond that anchors the C-terminal coil of the β-helix fold, which is expected to contribute to thermal stability only exists in the catalytic unit of Agl-ST.
- Department of Bioscience and Biotechnology, Fukui Prefectural University, 4-1-1 Matsuokakenjyoujima, Eiheiji-cho, Yoshida-gun, Fukui, 910-1195, Japan.
Organizational Affiliation: