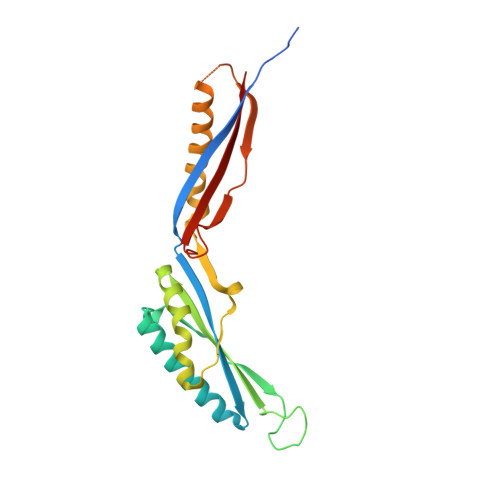
Structural analysis of a Simpl-like protein from Campylobacter jejuni.
Oh, H.B., Yoon, S.I.(2020) Biochem Biophys Res Commun 529: 270-276
- PubMed: 32703422
- DOI: https://doi.org/10.1016/j.bbrc.2020.05.211
- Primary Citation of Related Structures:
7C50, 7C51 - PubMed Abstract:
Signaling molecule that interacts with mouse pelle-like kinase (Simpl) is an animal protein that contributes to the regulation of inflammatory responses. Although Simpl-like proteins (SLPs) are mainly found in bacteria, functional and structural studies on bacterial SLPs are limited to BP26, a periplasmic protein from Brucella species. We identified a group of bacterial SLPs, including Campylobacter jejuni SLP (cjSLP) and Shewanella putrefaciens SLP (spSLP), that exhibit significant sequence variation from Simpl and BP26. To address the structural and oligomeric diversities of SLPs, we determined the crystal structure of cjSLP and performed a comparative analysis of SLP structures. cjSLP adopts a boomerang-shaped, two-domain structure, and each domain of cjSLP adopts an α-helix-decorated β-sheet structure as observed in BP26. This observation suggests that the duplicated α/β structure would be the canonical fold of the Simpl family. Despite the fold similarity, cjSLP exhibits a more open interdomain organization than BP26 and displays unique local structural features that are not observed in BP26. Furthermore, cjSLP and its ortholog spSLP are monomeric in solution in contrast to the hexadecameric assembly of BP26. Therefore, we conclude that cjSLP represents a unique bacterial SLP group that is distinct from BP26 in both structures and oligomeric states.
- Division of Biomedical Convergence, College of Biomedical Science, Kangwon National University, Chuncheon, 24341, Republic of Korea.
Organizational Affiliation: