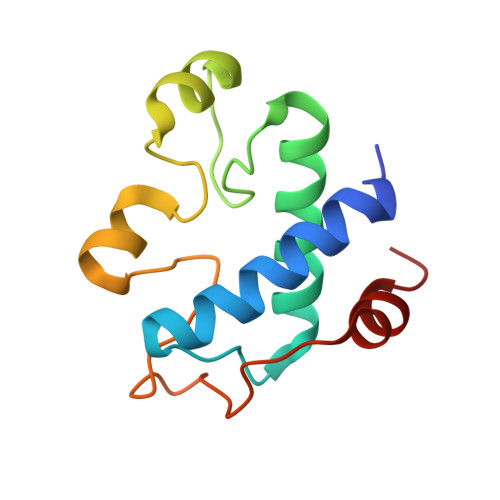
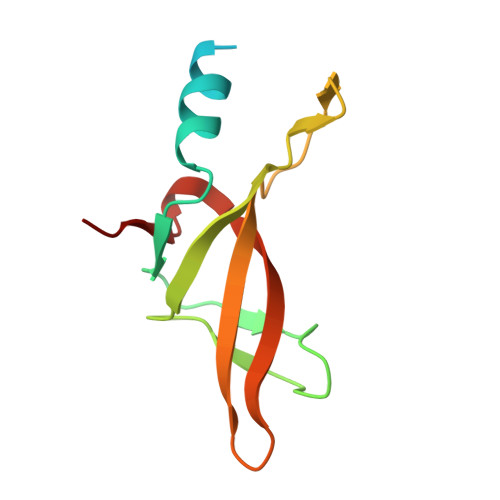
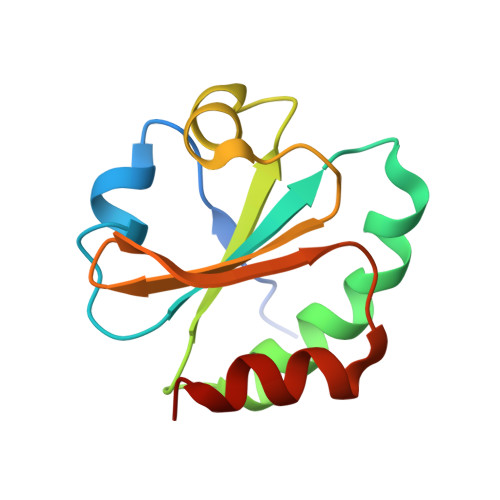
Structural basis for thioredoxin isoform-based fine-tuning of ferredoxin-thioredoxin reductase activity.
Juniar, L., Tanaka, H., Yoshida, K., Hisabori, T., Kurisu, G.(2020) Protein Sci 29: 2538-2545
- PubMed: 33015914
- DOI: https://doi.org/10.1002/pro.3964
- Primary Citation of Related Structures:
7BZK, 7C2B, 7C3F, 7C65 - PubMed Abstract:
Photosynthetic electron transport occurs on the thylakoid membrane of chloroplasts. Ferredoxin (Fd), the final acceptor in the electron transport chain, distributes electrons to several Fd-dependent enzymes including Fd-thioredoxin reductase (FTR). A cascade from Fd to FTR further reduces Thioredoxin (Trx), which tunes the activity of target metabolic enzymes eventually in a light-dependent manner. We previously reported that 10 Trx isoforms in Arabidopsis thaliana can be clustered into three classes based on the kinetics of the FTR-dependent reduction (high-, middle-, and low-efficiency classes). In this study, we determined the X-ray structure of three electron transfer complexes of FTR and Trx isoform, Trx-y1, Trx-f2, and Trx-m2, as representative examples of each class. Superposition of the FTR structure with/without Trx showed no main chain structural changes upon complex formation. There was no significant conformational change for single and complexed Trx-m structures. Nonetheless, the interface of FTR:Trx complexes displayed significant variation. Comparative analysis of the three structures showed two types of intermolecular interactions; (i) common interactions shared by all three complexes and (ii) isoform-specific interactions, which might be important for fine-tuning FTR:Trx activity. Differential electrostatic potentials of Trx isoforms may be key to isoform-specific interactions.
- Institute for Protein Research, Osaka University, Suita, Osaka, Japan.
Organizational Affiliation: